5B6V
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: resting state structure | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nango, E, Royant, A, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
6U2G
 
 | | BRAF-MEK complex with AMP-PCP bound to BRAF | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liau, N.P.D, Wendorff, T, Hymowitz, S, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7P3C
 
 | | EED in complex with compound 4 | | Descriptor: | N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, N-[5-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methylamino]-[1,2,4]triazolo[4,3-c]pyrimidin-8-yl]benzamide, Polycomb protein EED | | Authors: | Read, J.A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Diverse, Potent, and Efficacious Inhibitors That Target the EED Subunit of the Polycomb Repressive Complex 2 Methyltransferase.
J.Med.Chem., 64, 2021
|
|
7P3G
 
 | | EED in complex with compound 4 | | Descriptor: | N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, N5-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-N8-methyl-N8-(1-methylpyrazol-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidine-5,8-diamine, Polycomb protein EED | | Authors: | Read, J.A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Diverse, Potent, and Efficacious Inhibitors That Target the EED Subunit of the Polycomb Repressive Complex 2 Methyltransferase.
J.Med.Chem., 64, 2021
|
|
7P3J
 
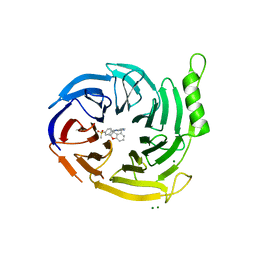 | | EED in complex with compound 4 | | Descriptor: | 8-[6-[(dimethylamino)methyl]-2-methyl-pyridin-3-yl]-5-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methylamino]-2H-pyrido[3,4-d]pyridazin-1-one, MAGNESIUM ION, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Read, J.A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Diverse, Potent, and Efficacious Inhibitors That Target the EED Subunit of the Polycomb Repressive Complex 2 Methyltransferase.
J.Med.Chem., 64, 2021
|
|
8F1Z
 
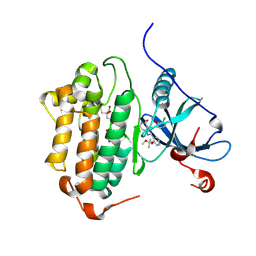 | | EGFR kinase in complex with Bayer #33 | | Descriptor: | (2P)-3-(3-chloro-2-methoxyanilino)-2-[3-(2-methoxy-2-methylpropoxy)pyridin-4-yl]-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, CITRIC ACID, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8F1Y
 
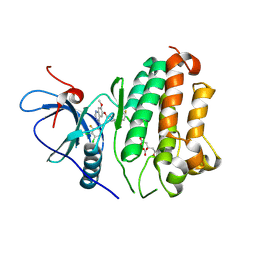 | | EGFR kinase in complex with poziotinib | | Descriptor: | 1-[4-[4-[[3,4-bis(chloranyl)-2-fluoranyl-phenyl]amino]-7-methoxy-quinazolin-6-yl]oxypiperidin-1-yl]propan-1-one, CITRIC ACID, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8F1H
 
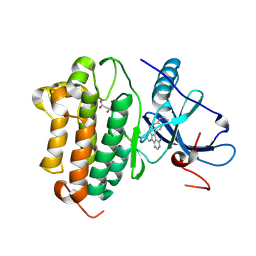 | | EGFR kinase in complex with TAS6417 (CLN-081) | | Descriptor: | CITRIC ACID, Epidermal growth factor receptor, N-[(5P,8S,10R)-4-amino-6-methyl-5-(quinolin-3-yl)-8,9-dihydropyrimido[5,4-b]indolizin-8-yl]prop-2-enamide | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-05 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8F1X
 
 | | EGFR kinase in complex with mobocertinib (TAK-788) | | Descriptor: | Epidermal growth factor receptor, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8F1W
 
 | | EGFR(T790M/V948R) kinase in complex with poziotinib | | Descriptor: | 1-[4-[4-[[3,4-bis(chloranyl)-2-fluoranyl-phenyl]amino]-7-methoxy-quinazolin-6-yl]oxypiperidin-1-yl]propan-1-one, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3OCZ
 
 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase Complexed with the inhibitor adenosine 5-O-thiomonophosphate | | Descriptor: | ADENOSINE -5'-THIO-MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of the inhibition of class C acid phosphatases by adenosine 5'-phosphorothioate.
Febs J., 278, 2011
|
|
7YVL
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | Descriptor: | Spike glycoprotein, TH272 Fab heavy chain, TH272 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
6TXR
 
 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5V3P
 
 | |
8PVQ
 
 | | Crystal Structure of Human PTX3 C-terminal domain | | Descriptor: | Pentraxin-related protein PTX3 | | Authors: | Levy, C.W. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Pentraxin 3 mediated crosslinking of heavy chain hyaluronan complexes: new insights from structural and biophysical studies
To Be Published
|
|
5HYX
 
 | | Plant peptide hormone receptor RGFR1 in complex with RGF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PTR-SER-ASN-PRO-GLY-HIS-HIS-PRO-HYP-ARG-HIS-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-01-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth
Cell Res., 26, 2016
|
|
8PWG
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 90-120 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Ortolani, G, Bosman, R, Branden, G, Neutze, R. | | Deposit date: | 2023-07-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWI
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 60-90 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Bosman, R, Ortolani, G, Branden, G, Neutze, R. | | Deposit date: | 2023-07-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWQ
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 120-150 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Bosman, R, Ortolani, G, Branden, G, Neutze, R. | | Deposit date: | 2023-07-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWJ
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography. 30-60 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Bosman, R, Ortolani, G, Branden, G, Neutze, R. | | Deposit date: | 2023-07-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWP
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 0-30 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Ortolani, G, Bosman, R, Branden, G, Neutze, R. | | Deposit date: | 2023-07-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
7YVN
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH281 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH281 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVE
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH027 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH027 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVP
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH272/281 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH272 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVG
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH132 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH132 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
