2LVP
 
 | | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
1TBU
 
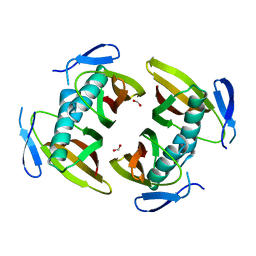 | |
1T77
 
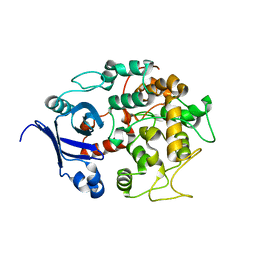 | | Crystal structure of the PH-BEACH domains of human LRBA/BGL | | Descriptor: | Lipopolysaccharide-responsive and beige-like anchor protein | | Authors: | Gebauer, D, Li, J, Jogl, G, Shen, Y, Myszka, D.G, Tong, L. | | Deposit date: | 2004-05-08 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the PH-BEACH Domains of Human LRBA/BGL
Biochemistry, 43, 2004
|
|
2LVN
 
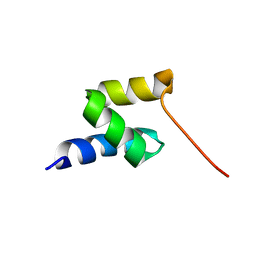 | | Structure of the gp78 CUE domain | | Descriptor: | E3 ubiquitin-protein ligase AMFR | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
4E14
 
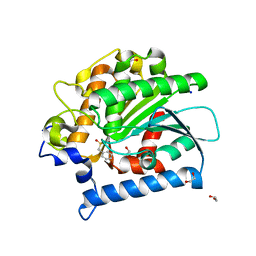 | | Crystal structure of kynurenine formamidase conjugated with phenylmethylsulfonyl fluoride | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, kynurenine formamidase | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2012-03-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster.
Biochem.J., 446, 2012
|
|
3KL4
 
 | | Recognition of a signal peptide by the signal recognition particle | | Descriptor: | Signal peptide of yeast dipeptidyl aminopeptidase B, Signal recognition 54 kDa protein | | Authors: | Janda, C.Y, Nagai, K, Li, J, Oubridge, C. | | Deposit date: | 2009-11-06 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Recognition of a signal peptide by the signal recognition particle.
Nature, 465, 2010
|
|
3L3Z
 
 | | Crystal structure of DHT-bound androgen receptor in complex with the third motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
5Y1Z
 
 | | Crystal structure of ZMYND8 PHD-BROMO-PWWP tandem in complex with Drebrin ADF-H domain | | Descriptor: | Drebrin, GLYCEROL, Protein kinase C-binding protein 1, ... | | Authors: | Yao, N, Li, J, Liu, H, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2017-07-22 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.676 Å) | | Cite: | The Structure of the ZMYND8/Drebrin Complex Suggests a Cytoplasmic Sequestering Mechanism of ZMYND8 by Drebrin
Structure, 25, 2017
|
|
4E11
 
 | | Crystal structure of kynurenine formamidase from Drosophila melanogaster | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2012-03-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster.
Biochem.J., 446, 2012
|
|
8YKW
 
 | | Cryo-EM structure of succinate receptor SUCR1 bound to succinic acid | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8YKV
 
 | | Cryo-EM structure of succinate receptor SUCR1 bound to compound 31 | | Descriptor: | (2~{S})-2-[[6-[4-(trifluoromethyloxy)phenyl]pyridin-2-yl]carbonylamino]butanedioic acid, Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8YKX
 
 | | Cryo-EM structure of succinate receptor SUCR1 bound to maleic acid | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
4E02
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with AMPPNP | | Descriptor: | (S)-2-chloro-3-phenylpropanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1547 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
4R8G
 
 | | Crystal Structure of Myosin-1c tail in complex with Calmodulin | | Descriptor: | Calmodulin, SULFATE ION, Unconventional myosin-Ic | | Authors: | Lu, Q, Li, J, Ye, F, Zhang, M. | | Deposit date: | 2014-09-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structure of myosin-1c tail bound to calmodulin provides insights into calcium-mediated conformational coupling.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4E15
 
 | |
7XAN
 
 | | Structure of a triple-helix region of human collagen type III from Trautec | | Descriptor: | Collagen alpha-1(III) chain | | Authors: | Qian, S, Li, H, Fan, X, Tian, X, Li, J, Wang, L, Chu, Y. | | Deposit date: | 2022-03-18 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a triple-helix region of human collagen type III from Trautec
To Be Published
|
|
6JRL
 
 | | Crystal structure of Drosophila alpha methyldopa-resistant protein/3,4-dihydroxyphenylacetaldehyde synthase | | Descriptor: | 3,4-dihydroxyphenylacetaldehyde synthase | | Authors: | Wei, S, Vavrick, C.J, Guan, H, Liao, C, Robinson, H, Liang, J, Wang, D, Han, Q, Li, J. | | Deposit date: | 2019-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the bifunctional mechanism of Drosophila alpha methyldopa-resistant protein/3,4-dihydroxyphenylacetaldehyde synthase
To Be Published
|
|
4FD6
 
 | |
4FD4
 
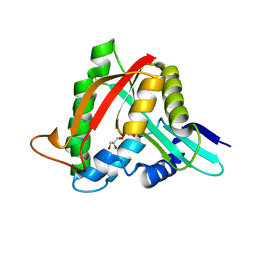 | |
7X8M
 
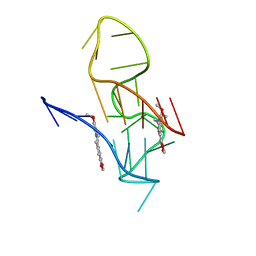 | | NMR Solution Structure of the 2:1 Berberine-KRAS-G4 Complex | | Descriptor: | BERBERINE, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8O
 
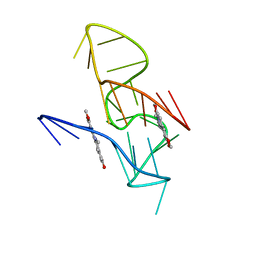 | | NMR Solution Structure of the 2:1 Coptisine-KRAS-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8N
 
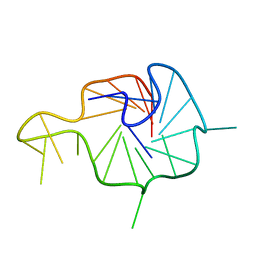 | | NMR Solution Structure of the Wild-type Bulge-containing KRAS-G4 | | Descriptor: | DNA (24-mer) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X2E
 
 | |
3L3X
 
 | | Crystal structure of DHT-bound androgen receptor in complex with the first motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
3UJL
 
 | | Crystal structure of abscisic acid bound PYL2 in complex with type 2C protein phosphatase ABI2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Soon, F.-F, Ng, L.-M, Kovach, A, Tan, M.H.E, Suino-Powell, K.M, He, Y, Xu, Y, Brunzelle, J.S, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases.
Science, 335, 2012
|
|
