6AQJ
 
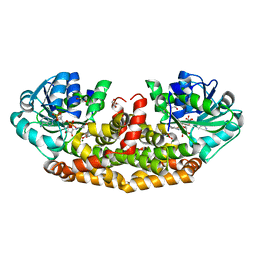 | | Crystal structures of Staphylococcus aureus ketol-acid reductoisomerase in complex with two transition state analogs that have biocidal activity. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Gracia, M, Lv, Y, Schembri, M.A, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-08-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.373 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
7XJJ
 
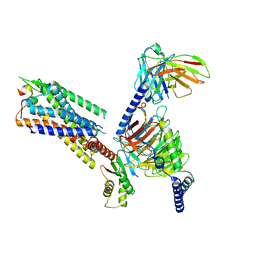 | | Cryo-EM structure of the galanin-bound GALR1-miniGo complex | | Descriptor: | G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Galanin, Galanin receptor type 1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8I79
 
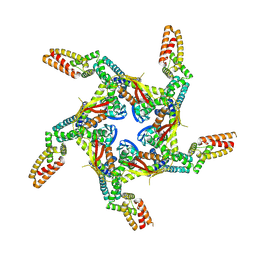 | | Cryo-EM structure of KCTD7 in complex with Cullin3 | | Descriptor: | BTB/POZ domain-containing protein KCTD7, Cullin-3 | | Authors: | Jiang, W, Wang, W, Zheng, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Sci Adv, 9, 2023
|
|
7X2D
 
 | | Cryo-EM structure of the tavapadon-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | 1,5-dimethyl-6-[2-methyl-4-[3-(trifluoromethyl)pyridin-2-yl]oxy-phenyl]pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-15 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7X2F
 
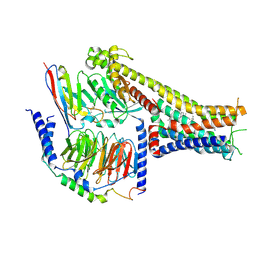 | | Cryo-EM structure of the dopamine and LY3154207-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7X2C
 
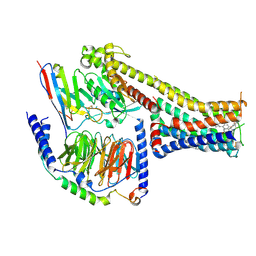 | | Cryo-EM structure of the fenoldopam-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
6KTR
 
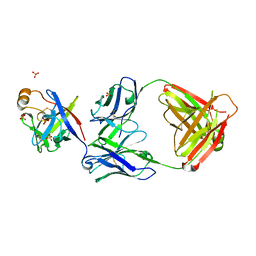 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
7XJL
 
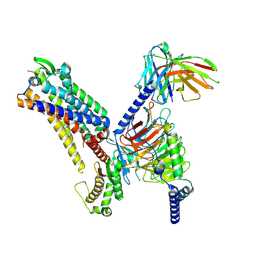 | | Cryo-EM structure of the spexin-bound GALR2-miniGq complex | | Descriptor: | Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XJK
 
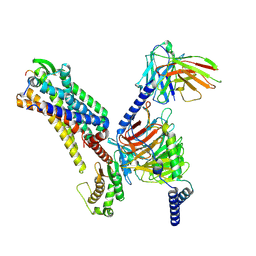 | |
7F24
 
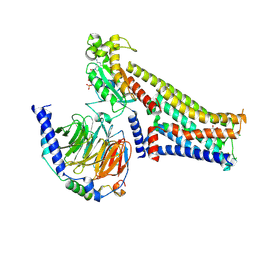 | |
7F0T
 
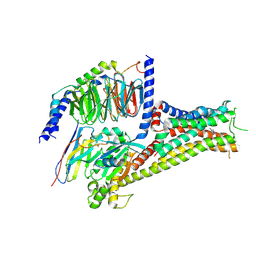 | |
7F23
 
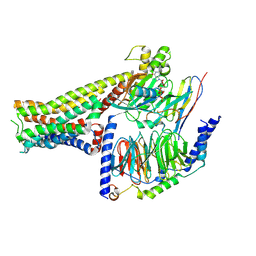 | |
7F1O
 
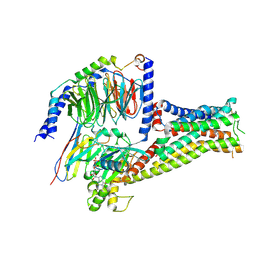 | |
7F1Z
 
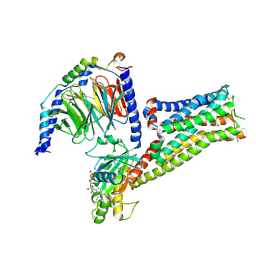 | |
2HV9
 
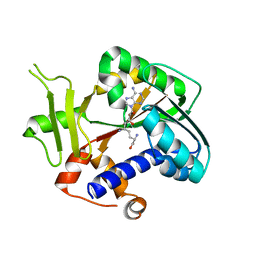 | | Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase in complex with sinefungin | | Descriptor: | SINEFUNGIN, mRNA cap guanine-N7 methyltransferase | | Authors: | Lima, C.D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Analysis of Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase, Structure of the Enzyme Bound to Sinefungin, and Evidence That Cap Methyltransferase Is the Target of Sinefungin's Antifungal Activity
J.Biol.Chem., 281, 2006
|
|
6IP0
 
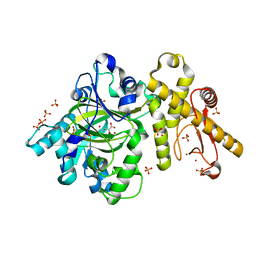 | |
6IP4
 
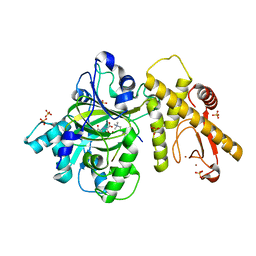 | |
2KHP
 
 | |
3KYT
 
 | |
3L0J
 
 | | Crystal structure of orphan nuclear receptor RORgamma in complex with natural ligand | | Descriptor: | (3alpha,8alpha,22R)-cholest-5-ene-3,22-diol, Nuclear receptor ROR-gamma, Nuclear receptor coactivator 2 | | Authors: | Martynowski, D, Li, Y. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma.
Mol.Endocrinol., 24, 2010
|
|
7XRQ
 
 | |
5WWO
 
 | | Crystal structure of Enp1 | | Descriptor: | Essential nuclear protein 1, Protein LTV1 | | Authors: | Ye, K, Zhang, W. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5WYJ
 
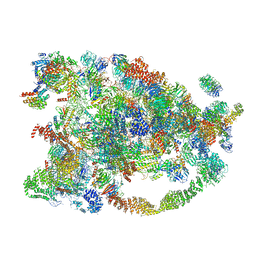 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WWN
 
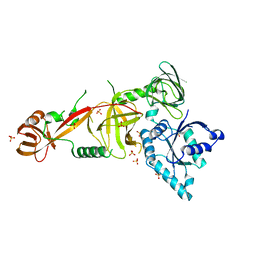 | | Crystal structure of Tsr1 | | Descriptor: | Ribosome biogenesis protein TSR1, SULFATE ION | | Authors: | Ye, K, Wang, B. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5WWM
 
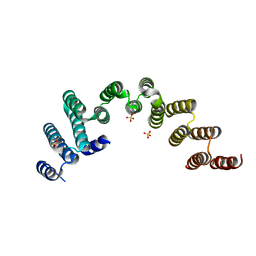 | | Crystal structure of the TPR domain of Rrp5 | | Descriptor: | SULFATE ION, rRNA biogenesis protein RRP5 | | Authors: | Ye, K, Chen, X. | | Deposit date: | 2017-01-03 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Correction: Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
