6KKA
 
 | | Xylanase J mutant from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, M, Takita, T, Nakatani, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
7C6A
 
 | | Crystal structure of AT2R-BRIL and SRP2070_Fab complex | | Descriptor: | IgG Light Chain, IgG heavy chain, SAR1, ... | | Authors: | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
7D7U
 
 | | Crystal structure of Ago2 MID domain in complex with 8-Br-adenosin-5'-monophosphate | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, Protein argonaute-2 | | Authors: | Suzuki, M, Takahashi, Y, Saito, J, Miyagi, H, Shinohara, F. | | Deposit date: | 2020-10-06 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | siRNA potency enhancement via chemical modifications of nucleotide bases at the 5'-end of the siRNA guide strand.
Rna, 27, 2021
|
|
3V66
 
 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH 2-(1-{2-[(4R,6S)-8-chloro-6-(2,3-dimethoxyphenyl)-4H,6H-pyrrolo[1,2-a][4,1]benzoxazepin-4-yl]acetyl}-4-piperidinyl)acetic acid | | Descriptor: | (1-{[(4R,6S)-8-chloro-6-(2,3-dimethoxyphenyl)-4H,6H-pyrrolo[1,2-a][4,1]benzoxazepin-4-yl]acetyl}piperidin-4-yl)acetic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Ohtsuka, M, Ohki, H, Haginoya, N, Itoh, M, Sugita, K, Usui, H, Ichikawa, M, Higashihashi, N. | | Deposit date: | 2011-12-18 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of novel tricyclic compounds as squalene synthase inhibitors
Bioorg.Med.Chem., 20, 2012
|
|
7CPK
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
7CPL
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
7C6B
 
 | | Crystal structure of Ago2 MID domain in complex with 6-(3-(2-carboxyethyl)phenyl)purine riboside monophosphate | | Descriptor: | 3-[3-[9-[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]purin-6-yl]phenyl]propanoic acid, PHOSPHATE ION, Protein argonaute-2 | | Authors: | Suzuki, M, Takahashi, Y, Saito, J, Miyagi, H, Shinohara, F. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | siRNA potency enhancement via chemical modifications of nucleotide bases at the 5'-end of the siRNA guide strand.
Rna, 27, 2021
|
|
3TK5
 
 | | Factor Xa in complex with D102-4380 | | Descriptor: | 4-{3-[(4-chlorophenyl)amino]-3-oxopropyl}-3-({[5-(propan-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]carbonyl}amino)benzoic acid, CALCIUM ION, Factor X heavy chain, ... | | Authors: | Suzuki, M, Mochizuki, A, Nagata, T, Takano, H, Kanno, H, Kishida, M, Ohta, T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zwitter ionic potent durable orally active Factor Xa inhibitor.
To be Published
|
|
3TK6
 
 | | factor Xa in complex with D46-5241 | | Descriptor: | CALCIUM ION, Factor X heavy chain, Factor X light chain, ... | | Authors: | Suzuki, M, Mochizuki, A, Nagata, T, Takano, H, Kanno, H, Kishida, M, Ohta, T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Zwitter ionic potent durable orally active Factor Xa inhibitor.
To be Published
|
|
4W4Q
 
 | | Glucose isomerase structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CALCIUM ION, Xylose isomerase | | Authors: | Nango, E, Tanaka, T, Sugahara, M, Suzuki, M, Iwata, S. | | Deposit date: | 2014-08-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat.Methods, 12, 2015
|
|
5D4J
 
 | | Chloride-bound form of a copper nitrite reductase from Alcaligenes faecals | | Descriptor: | ACETIC ACID, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4H
 
 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9UV5
 
 | | ligament intra-crystalline peptide (LICP) | | Descriptor: | PCA-PRO-ASP-HIS-GLU-GLY-THR-TYR-ASP-TYR | | Authors: | Futagawa, K, Suzuki, M. | | Deposit date: | 2025-05-09 | | Release date: | 2025-05-28 | | Method: | SOLUTION NMR | | Cite: | Elucidation of the aragonite nanofiber formation mechanism of LICP contained in hinge ligament of Pinctada fucata.
To Be Published
|
|
5GVO
 
 | | Solution NMR structure of a new lasso peptide sphaericin | | Descriptor: | Uncharacterized protein | | Authors: | Hemmi, H, Kodani, S, Inoue, Y, Suzuki, M, Dohra, H, Suzuki, T, Ohnishi-Kameyama, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Sphaericin, a Lasso Peptide from the Rare Actinomycete Planomonospora sphaerica
Eur.J.Org.Chem., 2017
|
|
3KLR
 
 | | Bovine H-protein at 0.88 angstrom resolution | | Descriptor: | GLYCEROL, Glycine cleavage system H protein, SULFATE ION | | Authors: | Higashiura, A, Kurakane, T, Matsuda, M, Suzuki, M, Inaka, K, Sato, M, Tanaka, H, Fujiwara, K, Nakagawa, A. | | Deposit date: | 2009-11-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | High-resolution X-ray crystal structure of bovine H-protein at 0.88 A resolution
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6LCQ
 
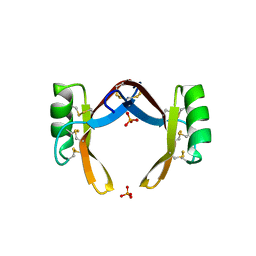 | | Crystal structure of rice defensin OsAFP1 | | Descriptor: | Defensin-like protein CAL1, PHOSPHATE ION | | Authors: | Ochiai, A, Ogawa, K, Fukuda, M, Suzuki, M, Ito, K, Tanaka, T, Sagehashi, Y, Taniguchi, M. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of rice defensin OsAFP1 and molecular insight into lipid-binding.
J.Biosci.Bioeng., 130, 2020
|
|
5KXV
 
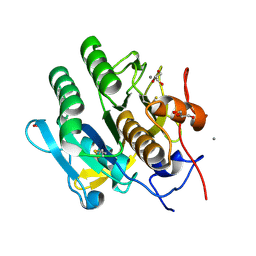 | | Structure Proteinase K at 0.98 Angstroms | | Descriptor: | CALCIUM ION, GLYCEROL, NITRATE ION, ... | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
5KXU
 
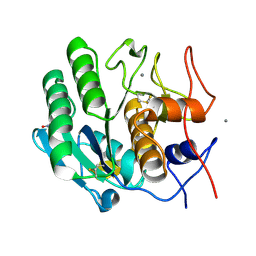 | | Structure Proteinase K determined by SACLA | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
3FI6
 
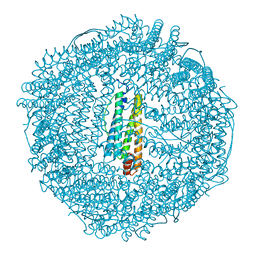 | | apo-H49AFr with high content of Pd ions | | Descriptor: | CADMIUM ION, Ferritin light chain, PALLADIUM ION, ... | | Authors: | Abe, M, Ueno, T, Hirata, K, Suzuki, M, Abe, S, Shimizu, N, Yamaoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2008-12-11 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Process of Accumulation of Metal Ions on the Interior Surface of apo-Ferritin: Crystal Structures of a Series of apo-Ferritins Containing Variable Quantities of Pd(II) Ions
J.Am.Chem.Soc., 131, 2009
|
|
3GCC
 
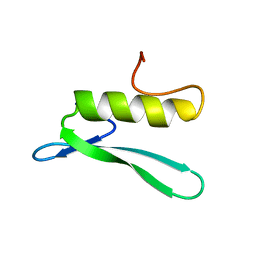 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, 46 STRUCTURES | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
5Y5N
 
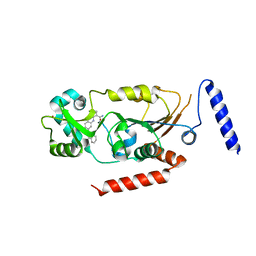 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | Deposit date: | 2017-08-09 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
7CE4
 
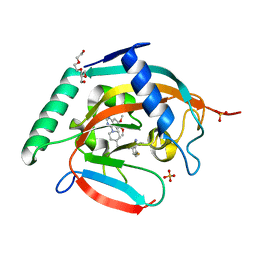 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
1O3Y
 
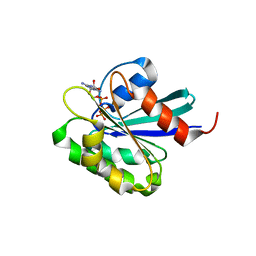 | | Crystal structure of mouse ARF1 (delta17-Q71L), GTP form | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
Nat.Struct.Biol., 10, 2003
|
|
1O3X
 
 | | Crystal structure of human GGA1 GAT domain | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-05-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Membrane Recruitment of Gga by Arf in Lysosomal Protein Transport
Nat.Struct.Biol., 10, 2003
|
|
2CZQ
 
 | | A novel cutinase-like protein from Cryptococcus sp. | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, cutinase-like protein | | Authors: | Masaki, K, Kamini, N.R, Ikeda, H, Iefuji, H, Kondo, H, Suzuki, M, Tsuda, S. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure and enhanced activity of a cutinase-like enzyme from Cryptococcus sp. strain S-2
Proteins, 77, 2009
|
|
