1OQN
 
 | | Crystal structure of the phosphotyrosine binding domain (PTB) of mouse Disabled 1 (Dab1) | | Descriptor: | Alzheimer's disease amyloid A4 protein homolog, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1 | | Authors: | Yun, M, Keshvara, L, Park, C.-G, Zhang, Y.-M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.-W. | | Deposit date: | 2003-03-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2
J.Biol.Chem., 278, 2003
|
|
1P3R
 
 | | Crystal structure of the phosphotyrosin binding domain(PTB) of mouse Disabled 1(Dab1) | | Descriptor: | Disabled homolog 2 | | Authors: | Yun, M, Keshvara, L, Park, C.G, Zhang, Y.M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.W. | | Deposit date: | 2003-04-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2.
J.Biol.Chem., 278, 2003
|
|
1PT9
 
 | | Crystal Structure Analysis of the DIII Component of Transhydrogenase with a Thio-Nicotinamide Nucleotide Analogue | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, GLYCEROL, NAD(P) transhydrogenase, ... | | Authors: | Singh, A, Venning, J.D, Quirk, P.G, van Boxel, G.I, Rodrigues, D.J, White, S.A, Jackson, J.B. | | Deposit date: | 2003-06-23 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Interactions between transhydrogenase and thio-nicotinamide analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation
J.Biol.Chem., 278, 2003
|
|
1PTJ
 
 | | Crystal structure analysis of the DI and DIII complex of transhydrogenase with a thio-nicotinamide nucleotide analogue | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Singh, A, Venning, J.D, Quirk, P.G, van Boxel, G.I, Rodrigues, D.J, White, S.A, Jackson, J.B. | | Deposit date: | 2003-06-23 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Interactions between transhydrogenase and thio-nicotinamide analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation
J.Biol.Chem., 278, 2003
|
|
1E3T
 
 | | Solution Structure of the NADP(H) binding Component (dIII) of Proton-Translocating Transhydrogenase from Rhodospirillum rubrum | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE (SUBUNIT BETA) | | Authors: | Jeeves, M, Smith, K.J, Quirk, P.G, Cotton, N.P.J, Jackson, J.B. | | Deposit date: | 2000-06-22 | | Release date: | 2000-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Nadp(H)-Binding Component (Diii) of Proton-Translocating Transhydrogenase from Rhodospirillum Rubrum
Biochim.Biophys.Acta, 1459, 2000
|
|
6YCK
 
 | | Crystal structure of GcoA T296A bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCL
 
 | | Crystal structure of GcoA T296G bound to p-vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCO
 
 | | Crystal structure of GcoA F169S bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCP
 
 | | Crystal structure of GcoA F169V bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCT
 
 | | Crystal structure of GcoA F169A_T296S bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Cytochrome P450, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-19 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCN
 
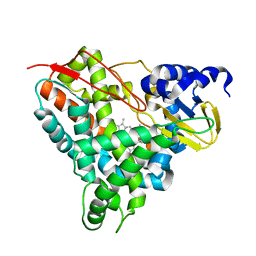 | | Crystal structure of GcoA F169A bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCI
 
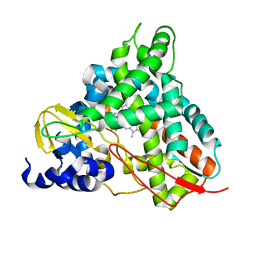 | | Crystal structure of GcoA T296G bound to guaiacol | | Descriptor: | Aromatic O-demethylase, cytochrome P450 subunit, Guaiacol, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
7R9X
 
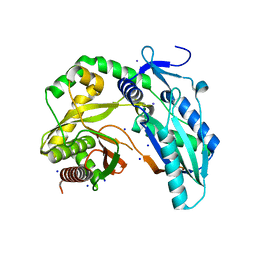 | |
1HBR
 
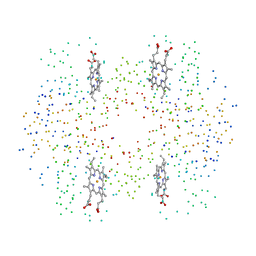 | | R-STATE FORM OF CHICKEN HEMOGLOBIN D | | Descriptor: | PROTEIN (HEMOGLOBIN D), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, E, Oliveira, M.A, Xie, Q, Ernst, S.R, Riggs, A.F, Hackert, M.L. | | Deposit date: | 1998-12-06 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural and functional analysis of the hemoglobin D component from chicken.
J.Biol.Chem., 274, 1999
|
|
6PBO
 
 | | Staphylococcus aureus Dihydrofolate reductase in complex with NADPH and UCP1232 | | Descriptor: | (4-{6-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-2H-1,3-benzodioxol-4-yl}phenyl)acetic acid, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Reeve, S.M, Wright, D.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Toward Broad Spectrum Dihydrofolate Reductase Inhibitors Targeting Trimethoprim Resistant Enzymes Identified in Clinical Isolates of Methicillin ResistantStaphylococcus aureus.
Acs Infect Dis., 5, 2019
|
|
6JJL
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-ARG-LYS-LEU, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy.
Commun Biol, 3, 2020
|
|
6JJK
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-TYR-LYS-ILE, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
6JJO
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | Periplasmic serine endoprotease DegP, TMB-CYRKL modulator | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.157 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
3SGY
 
 | |
3SH2
 
 | |
6P9Z
 
 | |
7SVJ
 
 | | Bile Salt Hydrolase from Lactobacillus ingluviei | | Descriptor: | CALCIUM ION, Choloylglycine hydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Walker, M.E, Patel, S, Redinbo, M.R. | | Deposit date: | 2021-11-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut.
Nat Microbiol, 8, 2023
|
|
7SVI
 
 | |
7SVH
 
 | | Bile Salt Hydrolase B from Lactobacillus gasseri | | Descriptor: | Choloylglycine hydrolase, MAGNESIUM ION | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2021-11-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut.
Nat Microbiol, 8, 2023
|
|
7SVG
 
 | |
