5IIE
 
 | |
7L06
 
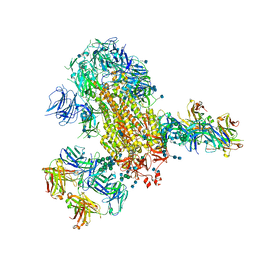 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L6M
 
 | |
7L6O
 
 | | Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 - gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2020-12-23 | | Release date: | 2021-04-14 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7LUA
 
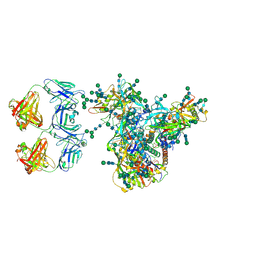 | | Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848 SOSIP gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-17 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7LU9
 
 | | Cryo-EM structure of DH851.3 bound to HIV-1 CH505 Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
5KG9
 
 | |
8DTK
 
 | |
7KDH
 
 | |
7KDI
 
 | |
7KEB
 
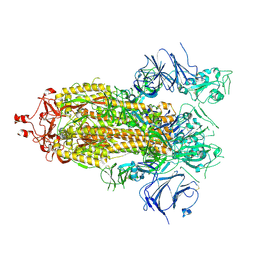 | |
7KEA
 
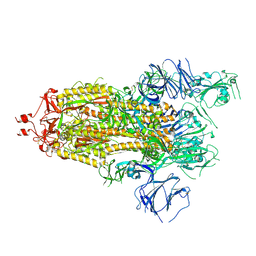 | |
7KDG
 
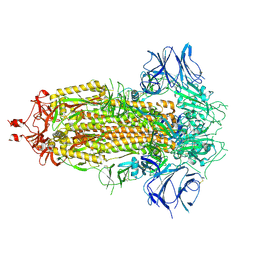 | |
7KE8
 
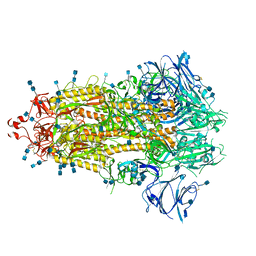 | |
7KDL
 
 | |
7KDK
 
 | |
8F5I
 
 | | SARS-CoV-2 S2 helix epitope scaffold bound by antibody DH1057.1 | | Descriptor: | DH1057.1 HC, DH1057.1 LC, Specialized acyl carrier protein, ... | | Authors: | Kapingidza, A.B, Wrapp, D, Winters, K, Azoitei, M.L. | | Deposit date: | 2022-11-14 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered immunogens to elicit antibodies against conserved coronavirus epitopes.
Nat Commun, 14, 2023
|
|
8F5H
 
 | |
8FDO
 
 | | SARS-CoV-2 fusion peptide epitope scaffold FP15 bound to DH1058 | | Descriptor: | DH1058 Heavy chain, DH1058 Light chain, FP15 | | Authors: | Kapingidza, A.B, Marston, D.J, Wrapp, D, Winters, K, Azoitei, M.L. | | Deposit date: | 2022-12-04 | | Release date: | 2023-10-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineered immunogens to elicit antibodies against conserved coronavirus epitopes.
Nat Commun, 14, 2023
|
|
7LD1
 
 | |
7LCN
 
 | |
7LAB
 
 | |
7LAA
 
 | |
7KE9
 
 | |
7KE4
 
 | |
