8DPW
 
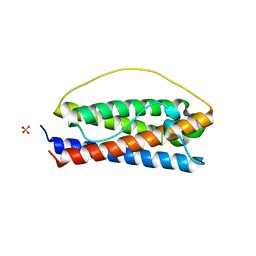 | | The structure of Interleukin-11 Mutein | | Descriptor: | Interleukin-11, SULFATE ION | | Authors: | Metcalfe, R.D, Griffin, M.D.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant.
Nat Commun, 14, 2023
|
|
8DPU
 
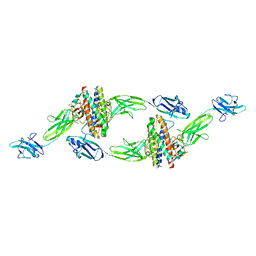 | | The crystal structure of the IL-11 signalling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Metcalfe, R.D, Aizel, K, Griffin, M.D.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant.
Nat Commun, 14, 2023
|
|
8DPS
 
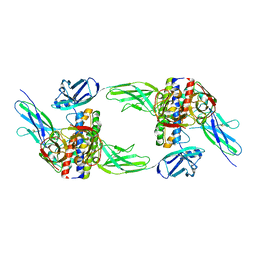 | | The structure of the interleukin 11 signalling complex, truncated gp130 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Metcalfe, R.D, Hanssen, E, Griffin, M.D.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant.
Nat Commun, 14, 2023
|
|
8DPT
 
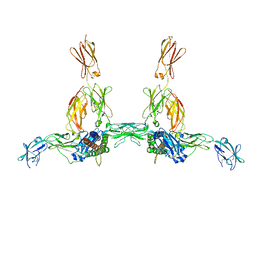 | | The structure of the IL-11 signalling complex, with full-length extracellular gp130 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Metcalfe, R.D, Hanssen, E, Griffin, M.D.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant.
Nat Commun, 14, 2023
|
|
6MUW
 
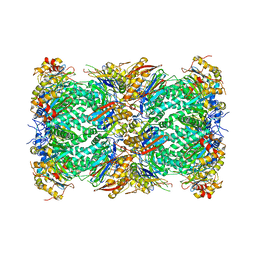 | | The structure of the Plasmodium falciparum 20S proteasome. | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Hanssen, E, Gillett, D.L, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2018-10-23 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
6MUV
 
 | | The structure of the Plasmodium falciparum 20S proteasome in complex with two PA28 activators | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Hanssen, E, Gillett, D.L, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2018-10-23 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
6MUX
 
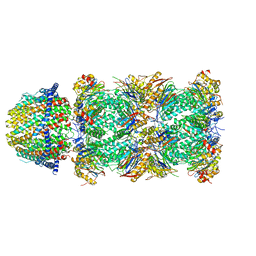 | | The structure of the Plasmodium falciparum 20S proteasome in complex with one PA28 activator | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Hanssen, E, Gillett, D.L, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2018-10-23 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
3CSJ
 
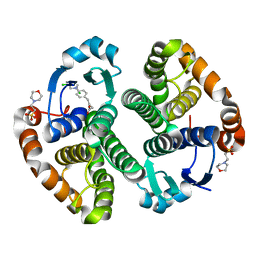 | | Human glutathione s-transferase p1-1 in complex with chlorambucil | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORAMBUCIL, CHLORIDE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The anti-cancer drug chlorambucil as a substrate for the human polymorphic enzyme glutathione transferase P1-1: kinetic properties and crystallographic characterisation of allelic variants.
J.Mol.Biol., 380, 2008
|
|
3CSI
 
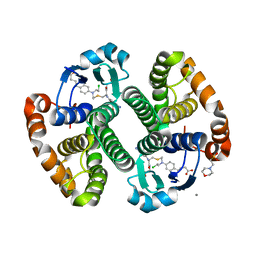 | |
3CSH
 
 | |
3DD3
 
 | |
3DGQ
 
 | |
2MBT
 
 | | NMR study of PaDsbA | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Rimmer, K, Mohanty, B, Scanlon, M.J. | | Deposit date: | 2013-08-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
3SL5
 
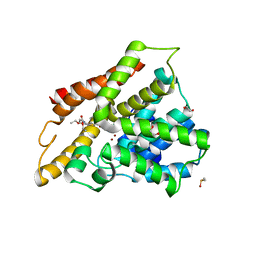 | | Crystal structure of the catalytic domain of PDE4D2 complexed with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6DFK
 
 | | Crystal structure of the 11S subunit of the Plasmodium falciparum proteasome, PA28 | | Descriptor: | SULFATE ION, Subunit of proteaseome activator complex,putative | | Authors: | Xie, S.C, Metcalfe, R.D, Gillett, D.L, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2018-05-15 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
3HKR
 
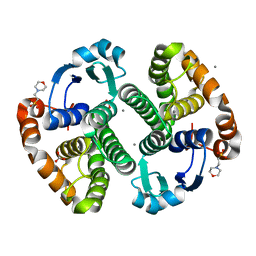 | | Crystal Structure of Glutathione Transferase Pi Y108V Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-05-25 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
3HJO
 
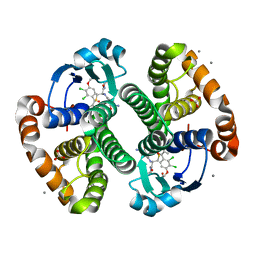 | |
3HJM
 
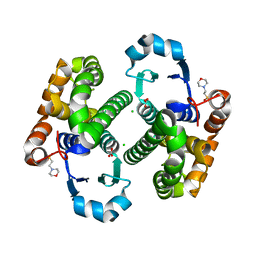 | | Crystal structure of human Glutathione Transferase Pi Y108V mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
3SL4
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10D | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL6
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 12c | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL8
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10o | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl 6-ethenyl 2-[(thiophen-2-ylacetyl)amino]-4,7-dihydrothieno[2,3-c]pyridine-3,6(5H)-dicarboxylate, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL3
 
 | | Crystal structure of the apo form of the catalytic domain of PDE4D2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5DIM
 
 | |
5DJL
 
 | |
5DCH
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I in complex with MIPS-0000851 (3-[(2-METHYLBENZYL)SULFANYL]-4H-1,2,4-TRIAZOL-4-AMINE) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, GLYCEROL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
