4ISB
 
 | |
4L1A
 
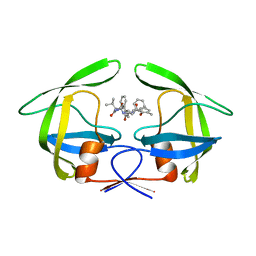 | | Crystallographic study of multi-drug resistant HIV-1 protease Lopinavir complex: mechanism of drug recognition and resistance | | Descriptor: | MDR769 HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE | | Authors: | Liu, Z, Yedidi, R.S, Wang, Y, Dewdney, T, Reiter, S, Brunzelle, J, Kovari, I, Kovari, L. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of multi-drug resistant HIV-1 protease lopinavir complex: mechanism of drug recognition and resistance.
Biochem.Biophys.Res.Commun., 437, 2013
|
|
6AO3
 
 | |
6AO4
 
 | |
4NN2
 
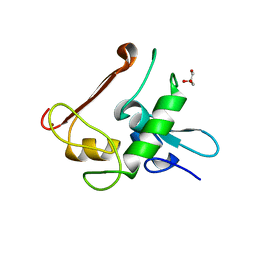 | | Protein Crystal Structure of Human Borjeson-Forssman-Lehmann Syndrome Associated Protein PHF6 | | Descriptor: | GLYCEROL, PHD finger protein 6, ZINC ION | | Authors: | Liu, Z, Li, F, Zhang, J, Mei, Y, Wu, J, Shi, Y. | | Deposit date: | 2013-11-16 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Crystal Structure of the second extended PHD domain of human PHF6 protein
J.Biol.Chem., 2014
|
|
4OU6
 
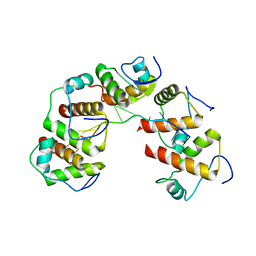 | | Crystal structure of DnaT84-153-dT10 ssDNA complex form 1 | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Primosomal protein 1 | | Authors: | Liu, Z, Chen, P, Niu, L, Teng, M, Li, X. | | Deposit date: | 2014-02-15 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode.
Nucleic Acids Res., 42, 2014
|
|
4OU7
 
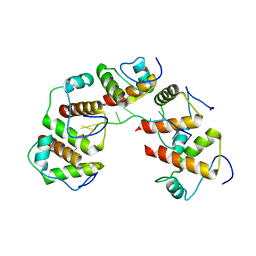 | | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Primosomal protein 1 | | Authors: | Liu, Z, Chen, P, Niu, L, Teng, M, Li, X. | | Deposit date: | 2014-02-15 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode.
Nucleic Acids Res., 42, 2014
|
|
8FZW
 
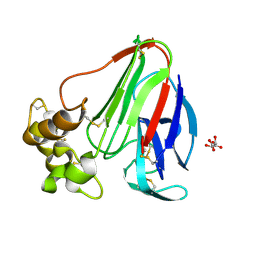 | | Thaumatin crystallized in cyclic olefin copolymer-based microfluidic chips | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Liu, Z, Gu, K, Shelby, M.L, Gilbile, D, Lyubimov, A.Y, Russi, S, Cohen, A.E, Coleman, M.A, Frank, M, Kuhl, T.L. | | Deposit date: | 2023-01-30 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A user-friendly plug-and-play cyclic olefin copolymer-based microfluidic chip for room-temperature, fixed-target serial crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6JUE
 
 | | The complex of PDZ and PBM | | Descriptor: | Partitioning defective 3 homolog, SULFATE ION, THR-ILE-ILE-THR-LEU | | Authors: | Liu, Z. | | Deposit date: | 2019-04-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Par complex cluster formation mediated by phase separation.
Nat Commun, 11, 2020
|
|
5ZAM
 
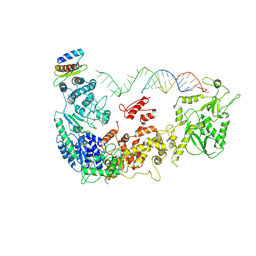 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2, RNA (73-mer) | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
5ZAK
 
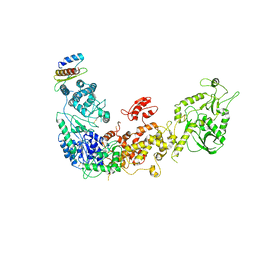 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
5ZAL
 
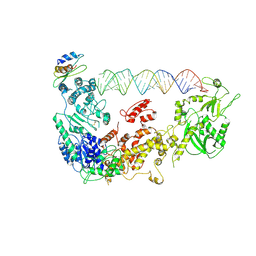 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2, RNA (73-mer) | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
4XH7
 
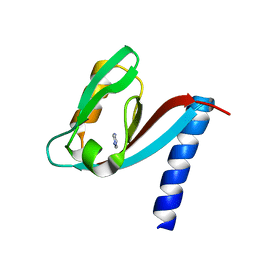 | | Crystal structure of MUPP1 PDZ4 | | Descriptor: | IMIDAZOLE, Multiple PDZ domain protein | | Authors: | Liu, Z, Zhu, H, Liu, W. | | Deposit date: | 2015-01-05 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and structural characterization of MUPP1-PDZ4 domain from Mus musculus.
Acta Biochim.Biophys.Sin., 47, 2015
|
|
3GBS
 
 | | Crystal structure of Aspergillus oryzae cutinase | | Descriptor: | Cutinase 1 | | Authors: | Gosser, Y, Lu, Z, Alemu, G, Li, H, Kong, X, Liu, Z, Montclare, J. | | Deposit date: | 2009-02-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation.
J.Am.Chem.Soc., 131, 2009
|
|
7UF8
 
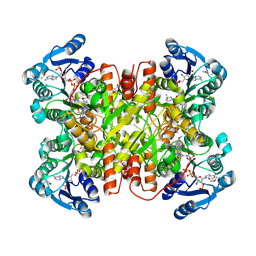 | | Structure of CtdP in complex with penicimutamide E and NADP+ | | Descriptor: | 1,2-ETHANEDIOL, CtdP, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Rivera, S, Liu, Z, Newmister, S.A, Gao, X, Sherman, D.H. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An NmrA-like enzyme-catalysed redox-mediated Diels-Alder cycloaddition with anti-selectivity.
Nat.Chem., 15, 2023
|
|
8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
3HEM
 
 | |
7EOD
 
 | | MITF HLHLZ Delta AKE | | Descriptor: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | Authors: | Li, P, Liu, Z, Fang, P, Wang, J. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7FD2
 
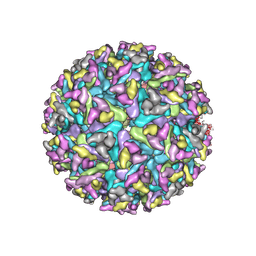 | | Cryo-EM structure of an alphavirus, Getah virus | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, Z, Liu, C, Wang, A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of infective Getah virus at 2.8 angstrom resolution determined by cryo-electron microscopy.
Cell Discov, 8, 2022
|
|
6UP4
 
 | |
4OZ6
 
 | | Structure of the Branched Intermediate in Protein Splicing | | Descriptor: | ALA-MET-ARG-TYR, MAGNESIUM ION, Mxe gyrA intein | | Authors: | Bick, M.J, Liu, Z, Frutos, S, Vila-Perello, M, Debelouchina, G.T, Darst, S.A, Muir, T.W. | | Deposit date: | 2014-02-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | Structure of the branched intermediate in protein splicing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7XCZ
 
 | |
7XDA
 
 | |
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | Authors: | Liu, Z, Liu, S, Gao, Y.Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
