6AEM
 
 | | Crystal structure of the PKD1 domain of Vibrio anguillarum Epp | | Descriptor: | CALCIUM ION, PKD domain, ZINC ION | | Authors: | Ma, Q, Li, P. | | Deposit date: | 2018-08-05 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | Structural basis for specific calcium binding by the polycystic-kidney-disease domain of Vibrio anguillarum protease Epp
Biochem. Biophys. Res. Commun., 505, 2018
|
|
2M0Q
 
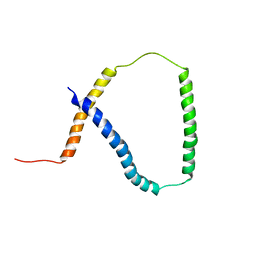 | | Solution NMR analysis of intact KCNE2 in detergent micelles demonstrate a straight transmembrane helix | | Descriptor: | Potassium voltage-gated channel subfamily E member 2 | | Authors: | Lai, C, Li, P, Chen, L, Zhang, L, Wu, F, Tian, C. | | Deposit date: | 2012-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential modulations of KCNQ1 by auxiliary proteins KCNE1 and KCNE2.
Sci Rep, 4, 2014
|
|
3U9J
 
 | |
3U9M
 
 | | Structure of reduced human FBXL5 hemerythrin like domain | | Descriptor: | F-box/LRR-repeat protein 5, FE (III) ION | | Authors: | Li, P. | | Deposit date: | 2011-10-19 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structural Basis of Iron Sensing by the Human F-box Protein FBXL5.
Chembiochem, 13, 2012
|
|
3TWY
 
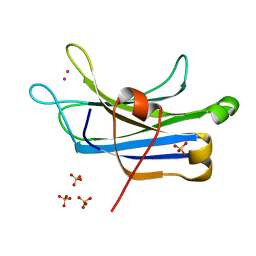 | | RAT PKC C2 DOMAIN BOUND TO PB | | Descriptor: | LEAD (II) ION, Protein kinase C alpha type, SULFATE ION | | Authors: | Li, P. | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pb2+ as Modulator of Protein-Membrane Interactions.
J.Am.Chem.Soc., 133, 2011
|
|
6IZC
 
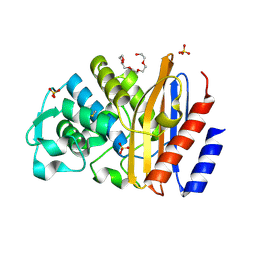 | |
6IZD
 
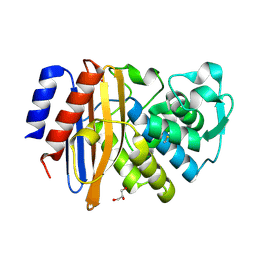 | |
3LRR
 
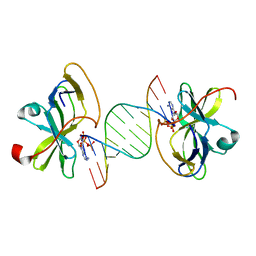 | | Crystal structure of human RIG-I CTD bound to a 12 bp AU rich 5' ppp dsRNA | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*(ATP)P*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*U)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Structure, 18, 2010
|
|
3LRN
 
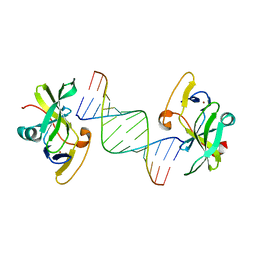 | | Crystal structure of human RIG-I CTD bound to a 14 bp GC 5' ppp dsRNA | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Structure, 18, 2010
|
|
5V5F
 
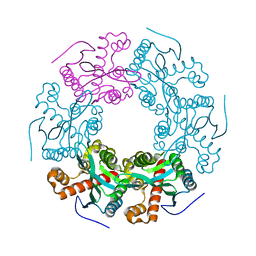 | | Crystal structure of RICE1 (PNT2) | | Descriptor: | At3g11770 | | Authors: | Li, P. | | Deposit date: | 2017-03-14 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | RISC-interacting clearing 3'- 5' exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana.
Elife, 6, 2017
|
|
3RDJ
 
 | | Rat PKC C2 domain Apo | | Descriptor: | Protein kinase C alpha type | | Authors: | Li, P. | | Deposit date: | 2011-04-01 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pb2+ as modulator of protein-membrane interactions.
J.Am.Chem.Soc., 133, 2011
|
|
4EMT
 
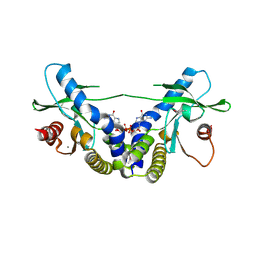 | | Crystal Structure of human STING bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Li, P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5JEJ
 
 | | Phosphorylated STING in complex with IRF-3 CTD | | Descriptor: | Interferon regulatory factor 3, Stimulator of interferon genes protein | | Authors: | Li, P, Shu, C. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3OG8
 
 | | Crystal structure of human RIG-I CTD bound to a 14-bp blunt-ended dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-08-16 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5' triphosphate.
Nucleic Acids Res., 39, 2011
|
|
4EMU
 
 | | Crystal structure of ligand free human STING | | Descriptor: | CALCIUM ION, Transmembrane protein 173 | | Authors: | Li, P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3S2X
 
 | | Structure of acetyl-Coenzyme A synthase Alpha subunit C-terminal domain | | Descriptor: | NICKEL (II) ION, acetyl-CoA synthase subunit alpha | | Authors: | Li, P. | | Deposit date: | 2011-05-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into the Mechanistic Role of the [Fe(4) S(4) ] Cubane in the A-Cluster {[Fe(4) S(4) ]-(SR)-[Ni(p) Ni(d) ]} of Acetyl-Coenzyme A Synthase.
Chembiochem, 12, 2011
|
|
4JL9
 
 | | Crystal structure of mouse TBK1 bound to BX795 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Li, P, Shu, C. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.0999 Å) | | Cite: | Structural Insights into the Functions of TBK1 in Innate Antimicrobial Immunity.
Structure, 21, 2013
|
|
4JLC
 
 | | Crystal structure of mouse TBK1 bound to SU6668 | | Descriptor: | 3-{2,4-dimethyl-5-[(Z)-(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-1H-pyrrol-3-yl}propanoic acid, Serine/threonine-protein kinase TBK1 | | Authors: | Li, P. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Functions of TBK1 in Innate Antimicrobial Immunity.
Structure, 21, 2013
|
|
7F6W
 
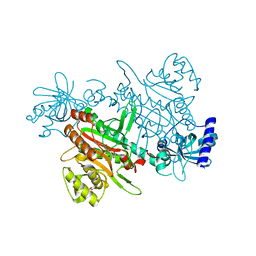 | | Crystal structure of Saccharomyces cerevisiae lysyl-tRNA Synthetase | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Lysine--tRNA ligase | | Authors: | Wu, S, Li, P, Hei, Z, Zheng, L, Wang, J, Fang, P. | | Deposit date: | 2021-06-26 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | Human lysyl-tRNA synthetase evolves a dynamic structure that can be stabilized by forming complex.
Cell.Mol.Life Sci., 79, 2022
|
|
5JEL
 
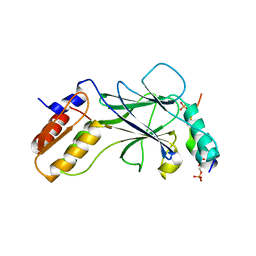 | | Phosphorylated TRIF in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, Phosphorylated TRIF peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEO
 
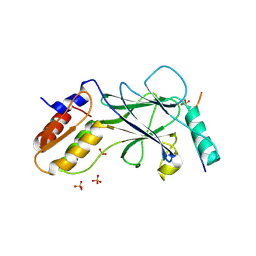 | | Phosphorylated Rotavirus NSP1 in complex with IRF-3 | | Descriptor: | Interferon regulatory factor 3, PHOSPHATE ION, Rotavirus NSP1 peptide | | Authors: | Zhao, B, Li, P. | | Deposit date: | 2016-04-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1NBF
 
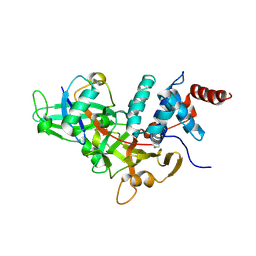 | | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde | | Descriptor: | Ubiquitin aldehyde, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Li, P, Li, M, Li, W, Yao, T, Wu, J.-W, Gu, W, Cohen, R.E, Shi, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-01-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde
Cell(Cambridge,Mass.), 111, 2002
|
|
1NB8
 
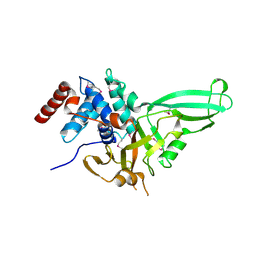 | | Structure of the catalytic domain of USP7 (HAUSP) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Li, P, Li, M, Li, W, Yao, T, Wu, J.-W, Gu, W, Cohen, R.E, Shi, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde
Cell(Cambridge,Mass.), 111, 2002
|
|
5C20
 
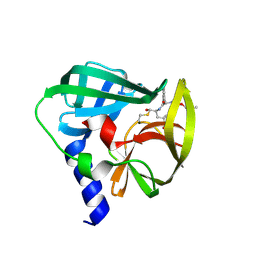 | | Crystal structure of EV71 3C Proteinase in complex with Compound 2 | | Descriptor: | 2-methylpropyl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1U
 
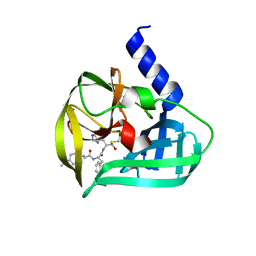 | | Crystal structure of EV71 3C Proteinase in complex with Compound Xb | | Descriptor: | (2S)-2-[[(E)-3-[4-(dimethylamino)phenyl]prop-2-enoyl]amino]-N-[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-phenyl-propanamide, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
