4XEY
 
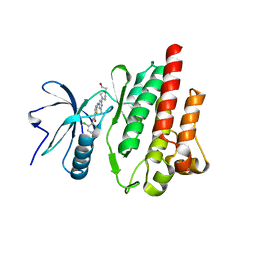 | |
4XUF
 
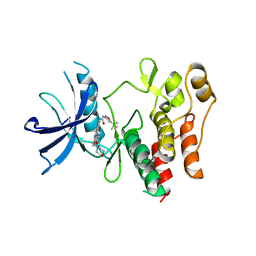 | | Crystal structure of the FLT3 kinase domain bound to the inhibitor quizartinib (AC220) | | Descriptor: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Zorn, J.A, Wang, Q, Fujimura, E, Barros, T, Kuriyan, J. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the FLT3 Kinase Domain Bound to the Inhibitor Quizartinib (AC220).
Plos One, 10, 2015
|
|
2RF9
 
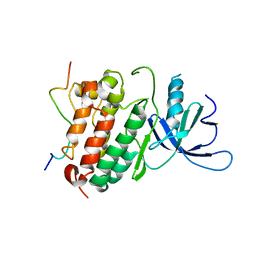 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RFE
 
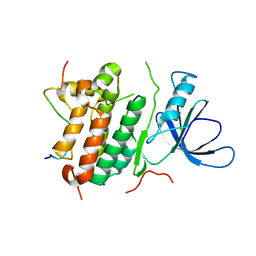 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RFD
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor, SULFATE ION | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
4Y95
 
 | | Crystal structure of the kinase domain of Bruton's tyrosine kinase with mutations in the activation loop | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, BETA-MERCAPTOETHANOL, ... | | Authors: | Wang, Q, Rosen, C.E, Kuriyan, J. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
4Y94
 
 | |
4Y93
 
 | | Crystal structure of the PH-TH-kinase construct of Bruton's tyrosine kinase (Btk) | | Descriptor: | 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, CALCIUM ION, Non-specific protein-tyrosine kinase,Non-specific protein-tyrosine kinase, ... | | Authors: | Wang, Q, Kuriyan, J. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
4U9G
 
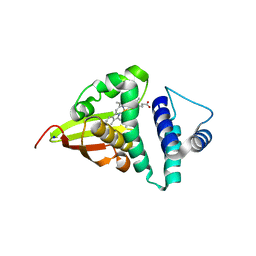 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)CO ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | CARBON MONOXIDE, NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9J
 
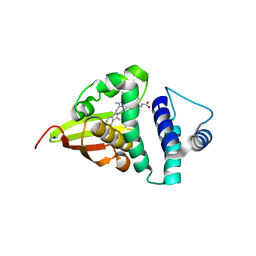 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, NO-binding heme-dependent sensor protein, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9B
 
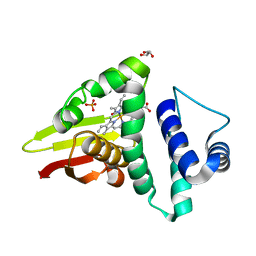 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)NO ligation state | | Descriptor: | GLYCEROL, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U9K
 
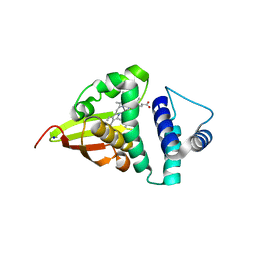 | | Crystal structure of an H-NOX protein from S. oneidensis in the Mn(II)NO ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | MANGANESE PROTOPORPHYRIN IX, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-06 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U99
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5BNB
 
 | |
6W69
 
 | | The structure of F64, S172A Keap1-BTB domain | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6W68
 
 | |
6W67
 
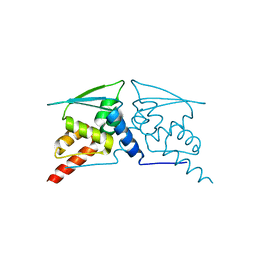 | |
6W66
 
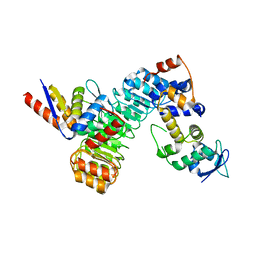 | | The structure of the F64A, S172A mutant Keap1-BTB domain in complex with SKP1-FBXL17 | | Descriptor: | F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, S-phase kinase-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6WCQ
 
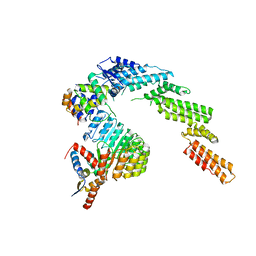 | | Structure of a substrate-bound DQC ubiquitin ligase | | Descriptor: | Cullin-1, F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, ... | | Authors: | Mena, E.L, Jevtic, P, Greber, B.J, Gee, C.L, Lew, B.G, Akopian, D, Nogales, E, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
1TDF
 
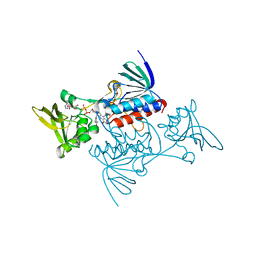 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THIOREDOXIN REDUCTASE REFINED AT 2 ANGSTROM RESOLUTION: IMPLICATIONS FOR A LARGE CONFORMATIONAL CHANGE DURING CATALYSIS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOREDOXIN REDUCTASE | | Authors: | Waksman, G, Krishna, T.S.R, Williams Junior, C.H, Kuriyan, J. | | Deposit date: | 1994-01-14 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Escherichia coli thioredoxin reductase refined at 2 A resolution. Implications for a large conformational change during catalysis.
J.Mol.Biol., 236, 1994
|
|
3KSY
 
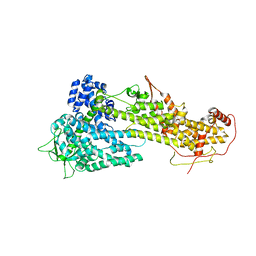 | |
4L9U
 
 | | Structure of C-terminal coiled coil of RasGRP1 | | Descriptor: | GLYCEROL, RAS guanyl-releasing protein 1, SULFATE ION | | Authors: | Iwig, J.S, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J.G, Wemmer, D.E, Roose, J.P, Kuriyan, J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6014 Å) | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
2OIQ
 
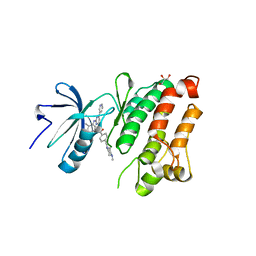 | | Crystal Structure of chicken c-Src kinase domain in complex with the cancer drug imatinib. | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Nagar, B, Frank, F, Cao, X, Henderson, M.N, Kuriyan, J. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | c-Src Binds to the Cancer Drug Imatinib with an Inactive Abl/c-Kit Conformation and a Distributed Thermodynamic Penalty.
Structure, 15, 2007
|
|
4L9M
 
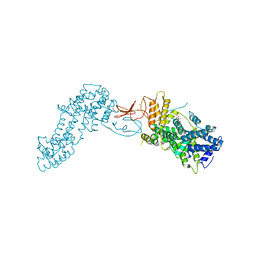 | | Autoinhibited state of the Ras-specific exchange factor RasGRP1 | | Descriptor: | CITRIC ACID, GLYCEROL, RAS guanyl-releasing protein 1, ... | | Authors: | Iwig, J.S, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J.G, Wemmer, D.E, Roose, J.P, Kuriyan, J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
4K2R
 
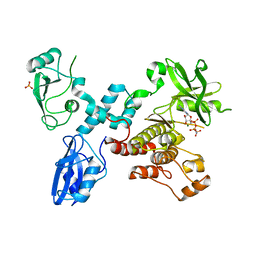 | | Structural basis for activation of ZAP-70 by phosphorylation of the SH2-kinase linker | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yan, Q, Barros, T, Visperas, P.R, Deindl, S, Kadlecek, T.A, Weiss, A, Kuriyan, J. | | Deposit date: | 2013-04-09 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Activation of ZAP-70 by Phosphorylation of the SH2-Kinase Linker.
Mol.Cell.Biol., 33, 2013
|
|
