7QND
 
 | | Cryo-EM structure of human full-length extrasynaptic beta3delta GABA(A)R in complex with THIP (gaboxadol), histamine and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5,6,7-tetrahydro-[1,2]oxazolo[5,4-c]pyridin-3-one, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QNA
 
 | | Cryo-EM structure of human full-length alpha4beta3gamma2 GABA(A)R in complex with GABA and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QNE
 
 | | Cryo-EM structure of human full-length synaptic alpha1beta3gamma2 GABA(A)R in complex with Ro15-4513 and megabody Mb38 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QNB
 
 | | Cryo-EM structure of human full-length beta3gamma2 GABA(A)R in complex with GABA and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QN8
 
 | | Cryo-EM structure of human full-length beta3delta GABA(A)R in complex with histamine and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7PCH
 
 | | Human carboxyhemoglobin bound to Staphylococcus aureus hemophore IsdB - 1:2 complex | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PCQ
 
 | | Human carboxyhemoglobin bound to Staphylococcus aureus hemophore IsdB - 1:1 complex | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PCF
 
 | | Human methemoglobin bound to Staphylococcus aureus hemophore IsdB | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.82 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
7A4M
 
 | | Cryo-EM structure of mouse heavy-chain apoferritin at 1.22 A | | Descriptor: | FE (III) ION, Ferritin heavy chain, ZINC ION | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (1.22 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
8BHI
 
 | | GABA-A receptor a5 homomer - a5V3 - RO5211223 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, [1,1-bis(oxidanylidene)-1,4-thiazinan-4-yl]-[6-[[5-methyl-3-(6-methylpyridin-3-yl)-1,2-oxazol-4-yl]methoxy]pyridin-3-yl]methanone | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BEJ
 
 | | GABA-A receptor a5 homomer - a5V3 - APO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHM
 
 | | GABA-A receptor a5 homomer - a5V3 - DMCM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, methyl 4-ethyl-6,7-dimethoxy-9H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHO
 
 | | GABA-A receptor a5 homomer - a5V3 - L655708 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, ethyl (7~{S})-15-methoxy-12-oxidanylidene-2,4,11-triazatetracyclo[11.4.0.0^{2,6}.0^{7,11}]heptadeca-1(17),3,5,13,15-pentaene-5-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHQ
 
 | | GABA-A receptor a5 homomer - a5V3 - RO7172670 | | Descriptor: | 2-[[5-methyl-3-(6-methylpyridazin-3-yl)-1,2-oxazol-4-yl]methyl]-5-(5-oxa-2-azaspiro[3.5]nonan-2-yl)pyridazin-3-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Kasaragod, V.B. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHS
 
 | | GABA-A receptor a5 homomer - a5V3 - RO4938581 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[bis(fluoranyl)methyl]-15-bromanyl-2,4,8,9,11-pentazatetracyclo[11.4.0.0^{2,6}.0^{8,12}]heptadeca-1(13),3,5,9,11,14,16-heptaene, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHB
 
 | | GABA-A receptor a5 homomer - a5V3 - RO154513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, ethyl 8-[(azanylidene-$l^{4}-azanylidene)amino]-5-methyl-6-oxidanylidene-4~{H}-imidazo[1,5-a][1,4]benzodiazepine-3-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y, Wahid, A.A. | | Deposit date: | 2022-10-30 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHA
 
 | | GABA-A receptor a5 homomer - a5V3 - Basmisanil - HR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Basmisanil, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Malinauskas, T.M, Wahid, A.A, Hardwick, S.W, Chirgadze, D.Y, Miller, P.S. | | Deposit date: | 2022-10-30 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4OXP
 
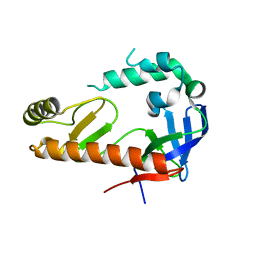 | |
8B0I
 
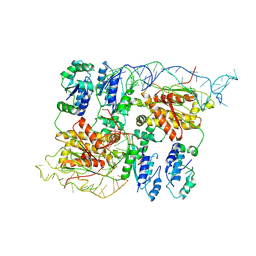 | | CryoEM structure of bacterial RapZ.GlmZ complex central to the control of cell envelope biogenesis | | Descriptor: | GlmZ small regulatory RNA, RNase adapter protein RapZ | | Authors: | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-10-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
8B0J
 
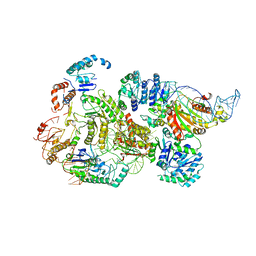 | | CryoEM structure of bacterial RNaseE.RapZ.GlmZ complex central to the control of cell envelope biogenesis | | Descriptor: | GlmZ small RNA, RNase adapter protein RapZ, Ribonuclease E | | Authors: | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-10-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
7A8Z
 
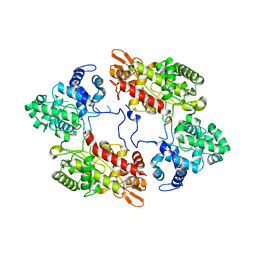 | |
7AA3
 
 | |
5NB9
 
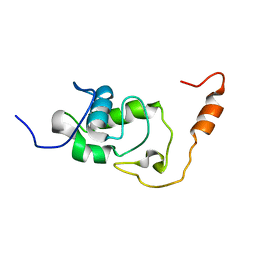 | | Structure of the N-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
5NBB
 
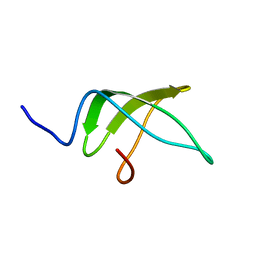 | | Structure of the C-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
