5Z9X
 
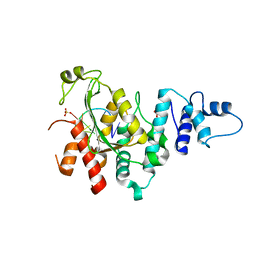 | | Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 in complex with an RNA substrate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*GP*CP*CP*CP*AP*UP*UP*AP*G)-3'), SULFATE ION, ... | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Gan, J, Cao, C, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
5Z9Z
 
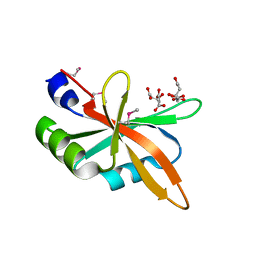 | | The C-terminal RRM domain of Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 (E329A/E330A/E332A) | | Descriptor: | CITRATE ANION, Small RNA degrading nuclease 1 | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Cao, C, Gan, J, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
1L6R
 
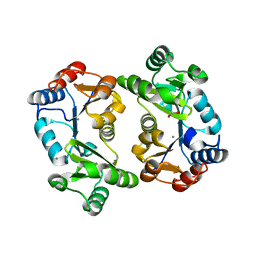 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC0014) | | Descriptor: | CALCIUM ION, FORMIC ACID, HYPOTHETICAL PROTEIN TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A.M, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-03-13 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum.
J.Biol.Chem., 279, 2004
|
|
1NOG
 
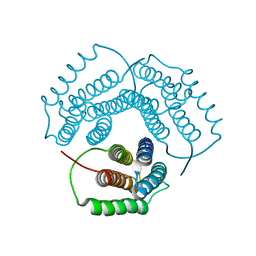 | | Crystal Structure of Conserved Protein 0546 from Thermoplasma Acidophilum | | Descriptor: | conserved hypothetical protein TA0546 | | Authors: | Saridakis, V, Sanishvili, R, Iakounine, A, Xu, X, Pennycooke, M, Gu, J, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for methylmalonic aciduria. The crystal structure of archaeal ATP:cobalamin adenosyltransferase.
J.Biol.Chem., 279, 2004
|
|
5VBN
 
 | | Crystal Structure of human DNA polymerase epsilon B-subunit in complex with C-terminal domain of catalytic subunit | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit 2, SULFATE ION, ... | | Authors: | Baranovskiy, A.G, Gu, J, Suwa, Y, Babayeva, N.D, Tahirov, T.H. | | Deposit date: | 2017-03-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the human Pol B-subunit in complex with the C-terminal domain of the catalytic subunit.
J. Biol. Chem., 292, 2017
|
|
1QAI
 
 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), MERCURY (II) ION, REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
2G3A
 
 | | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens | | Descriptor: | acetyltransferase | | Authors: | Cymborowski, M, Xu, X, Chruszcz, M, Zheng, H, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2FSR
 
 | | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, acetyltransferase | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Gu, J, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58
To be Published
|
|
2G7S
 
 | | The crystal structure of transcriptional regulator, TetR family, from Agrobacterium tumefaciens | | Descriptor: | transcriptional regulator, TetR family | | Authors: | Lunin, V.V, Chang, C, Xu, X, Gu, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structure of transcriptional regulator, TetR family, from Agrobacterium tumefaciens
To be Published
|
|
2GMY
 
 | | Crystal Structure of a Protein of Unknown Function ATU0492 from Agrobacterium tumefaciens, Putative Antioxidant Defence Protein AhpD | | Descriptor: | Hypothetical protein Atu0492 | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a hypothetical protein Atu0492 from Agrobacterium tumefaciens
To be Published
|
|
2FEX
 
 | | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, conserved hypothetical protein | | Authors: | Cymborowski, M.T, Wang, S, Chruszcz, M, Shumilin, I, Gu, J, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens
To be Published
|
|
2H8O
 
 | | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens | | Descriptor: | Geranyltranstransferase | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2IAI
 
 | | Crystal structure of SCO3833, a member of the TetR transcriptional regulator family from Streptomyces coelicolor A3 | | Descriptor: | Putative transcriptional regulator SCO3833 | | Authors: | Zimmerman, M.D, Xu, X, Wang, S, Gu, J, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of SCO3833, a member of the TetR transcriptional regulator family from Streptomyces coelicolor A3
To be Published
|
|
4HJJ
 
 | | Structure Reveals Function of the Dual Variable Domain Immunoglobulin (DVD-Ig) Molecule | | Descriptor: | Anti-IL12 Anti-IL18 DFab Heavy Chain, Anti-IL12 Anti-IL18 DFab Light Chain, CHLORIDE ION, ... | | Authors: | Jakob, C.G, Edalji, R, Judge, R.A, DiGiammarino, E, Li, Y, Gu, J, Ghayur, T. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure reveals function of the dual variable domain immunoglobulin (DVD-Ig[TM]) molecule
MAbs, 5, 2013
|
|
4IIO
 
 | | Crystal Structure of the Second SH3 Domain of ITSN2 Bound with a Synthetic Peptide | | Descriptor: | CHLORIDE ION, Intersectin-2, SULFATE ION, ... | | Authors: | Dong, A, Guan, X, Huang, H, Gu, J, Tempel, W, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Second SH3 Domain of ITSN2 Bound with a Synthetic Peptide
TO BE PUBLISHED
|
|
4IIM
 
 | | Crystal structure of the Second SH3 Domain of ITSN1 bound with a synthetic peptide | | Descriptor: | Intersectin-1, UNKNOWN ATOM OR ION, peptide ligand | | Authors: | Dong, A, Guan, X, Huang, H, Wernimont, A, Gu, J, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Second SH3 Domain of ITSN1 bound with a synthetic peptide
To be Published
|
|
2NWH
 
 | | Carbohydrate kinase from Agrobacterium tumefaciens | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Osipiuk, J, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | X-ray crystal structure of carbohydrate kinase from Agrobacterium tumefaciens
To be Published
|
|
4GLM
 
 | | Crystal structure of the SH3 Domain of DNMBP protein [Homo sapiens] | | Descriptor: | Dynamin-binding protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Guan, X, Huang, H, Tempel, W, Gu, J, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-14 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SH3 Domain of DNMBP protein [Homo sapiens]
to be published
|
|
4RR2
 
 | | Crystal structure of human primase | | Descriptor: | DNA primase large subunit, DNA primase small subunit, IRON/SULFUR CLUSTER, ... | | Authors: | Baranovskiy, A.G, Gu, J, Suwa, Y, Babayeva, N.D, Tahirov, T.H. | | Deposit date: | 2014-11-05 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the human primase.
J.Biol.Chem., 290, 2015
|
|
4Q5V
 
 | | Crystal structure of the catalytic core of human DNA polymerase alpha in ternary complex with an RNA-primed DNA template and aphidicolin | | Descriptor: | (3R,4R,4aR,6aS,8R,9R,11aS,11bS)-4,9-bis(hydroxymethyl)-4,11b-dimethyltetradecahydro-8,11a-methanocyclohepta[a]naphthale ne-3,9-diol, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Baranovskiy, A.G, Babayeva, N.D, Suwa, Y, Gu, J, Tahirov, T.H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for inhibition of DNA replication by aphidicolin.
Nucleic Acids Res., 42, 2014
|
|
5J5N
 
 | |
5J4U
 
 | |
7VYV
 
 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural biology and functional features of phage-derived depolymerase Depo32 on Klebsiella pneumoniae with K2 serotype capsular polysaccharides.
Microbiol Spectr, 11, 2023
|
|
7VZ3
 
 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural biology and functional features of phage-derived depolymerase Depo32 on Klebsiella pneumoniae with K2 serotype capsular polysaccharides.
Microbiol Spectr, 11, 2023
|
|
6JFW
 
 | | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket | | Descriptor: | PA0833-PD protein | | Authors: | Lin, X, Ye, F, Lin, S, Yang, F.L, Chen, Z.M, Cao, Y, Chen, Z.J, Gu, J, Lu, G.W. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
