9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
9BBC
 
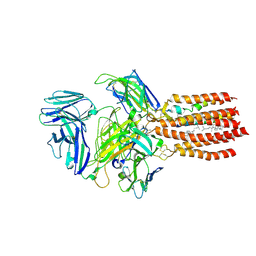 | | TCR GDN detergent micelle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2024-04-05 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
9C3E
 
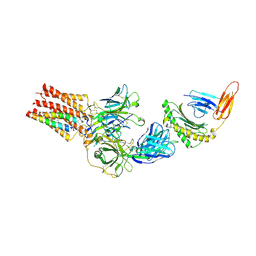 | | TCR - CD3 complex bound to HLA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Cancer/testis antigen 1,Beta-2-microglobulin,MHC class I antigen, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2024-05-31 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
6M0T
 
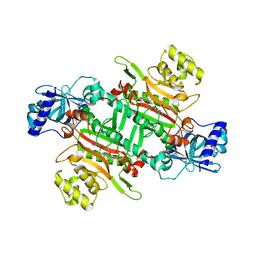 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin derivative (CL-2) | | Descriptor: | (3R)-3-[(R)-[(2R,6S)-6-methyloxan-2-yl]-oxidanyl-methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y. | | Deposit date: | 2020-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Cladosporin-Based Inhibitors of Malaria Parasites.
Acs Infect Dis., 7, 2021
|
|
7Z0X
 
 | | THSC20.HVTR26 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
7Z0Y
 
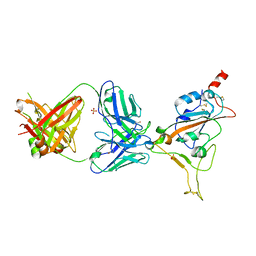 | | THSC20.HVTR04 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
8DBB
 
 | | Crystal structure of DDT with the selective inhibitor 2,5-Pyridinedicarboxylic Acid | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase, pyridine-2,5-dicarboxylic acid | | Authors: | Parkins, A, Banumathi, S, Pantouris, G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 2,5-Pyridinedicarboxylic acid is a bioactive and highly selective inhibitor of D-dopachrome tautomerase.
Structure, 31, 2023
|
|
8TW4
 
 | | TCR in nanodisc ND-I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2023-08-20 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
8TW6
 
 | | TCR in nanodisc ND-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, T cell receptor beta variable 6-5,T cell receptor beta chain MC.7.G5,MCHERRY, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2023-08-20 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
6HRK
 
 | | Structure of a far-red fluorescent biliprotein derived from a far-red induced allophycocyanin F subunit from a thermophilic cyanobacterium Chroococcidiopsis thermalis | | Descriptor: | Allophycocyanin beta-18 subunit apoprotein, BILIVERDINE IX ALPHA | | Authors: | Hou, Y.-N, Hoeppner, A, Ding, W.-L, Gaertner, W, Zhao, K.-H. | | Deposit date: | 2018-09-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Control of a far-red/near-infrared spectral switch in an artificial fluorescent biliprotein derived from allophycocyanin
Protein Sci., Suppl.: Diskette Appendix To V. , No. , [Month], Filename:, 31, 2022
|
|
6K3X
 
 | |
6K3Y
 
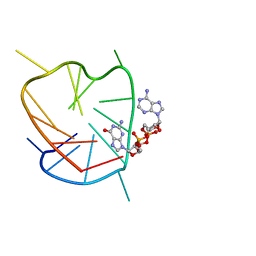 | | G-quadruplex complex with cyclic dinucleotide 3'-3' cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, DNA (5'-D(*TP*TP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*T)-3') | | Authors: | Winnerdy, F.R, Heddi, B, Phan, A.T. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a G-Quadruplex Bound to Linear- and Cyclic-Dinucleotides.
J.Am.Chem.Soc., 141, 2019
|
|
6XMX
 
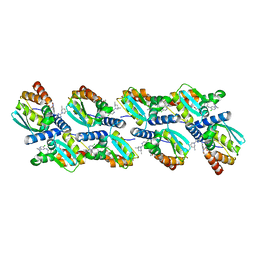 | | Cryo-EM structure of BCL6 bound to BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Yoon, H, Burman, S.S.R, Hunkeler, M, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2020-07-01 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Small-molecule-induced polymerization triggers degradation of BCL6.
Nature, 588, 2020
|
|
8SMU
 
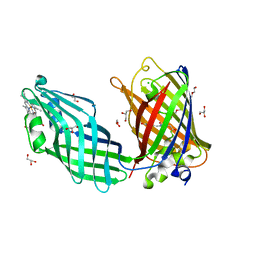 | | Integral fusion of the HtaA CR2 domain from Corynebacterium diphtheriae within EGFP | | Descriptor: | CHLORIDE ION, GLYCEROL, HtaACR2 integral fusion within enhanced green fluorescent protein, ... | | Authors: | Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development and atomic structure of a new fluorescence-based sensor to probe heme transfer in bacterial pathogens.
J.Inorg.Biochem., 249, 2023
|
|
6D2Q
 
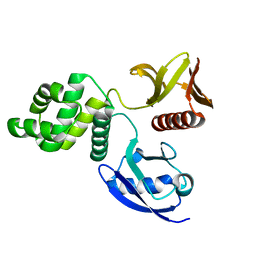 | | Crystal structure of the FERM domain of zebrafish FARP1 | | Descriptor: | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | | Authors: | Kuo, Y.C, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
6D21
 
 | |
6D2K
 
 | | Crystal structure of the FERM domain of mouse FARP2 | | Descriptor: | FERM, ARHGEF and pleckstrin domain-containing protein 2 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
7C2P
 
 | | Structure of Egk Peptide | | Descriptor: | Plant defensing Egk | | Authors: | El Sahili, A. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modulation of Lymphocyte Potassium Channel KV1.3 by Membrane-Penetrating, Joint-Targeting Immunomodulatory Plant Defensin.
Acs Pharmacol Transl Sci, 3, 2020
|
|
7C31
 
 | | Crystal structure of the grapevine defensin VvK1 | | Descriptor: | Knot1 domain-containing protein | | Authors: | Chen, M.W, Chang, S.C, Chandy, K.G, Luo, D. | | Deposit date: | 2020-05-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Modulation of Lymphocyte Potassium Channel KV1.3 by Membrane-Penetrating, Joint-Targeting Immunomodulatory Plant Defensin.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6LD0
 
 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with C6-hexyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-hexyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6ICE
 
 | | Crystal structure of Hamster MIF | | Descriptor: | Macrophage migration inhibitory factor | | Authors: | Sundaram, R, Vasudevan, D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macrophage migration inhibitory factor of Syrian golden hamster shares structural and functional similarity with human counterpart and promotes pancreatic cancer.
Sci Rep, 9, 2019
|
|
6LDB
 
 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with uronic isofagomine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LD6
 
 | |
6LEM
 
 | | Structure of E. coli beta-glucuronidase complex with C6-nonyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-nonyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LEJ
 
 | | Structure of E. coli beta-glucuronidase complex with C6-propyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.617 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
