3MVW
 
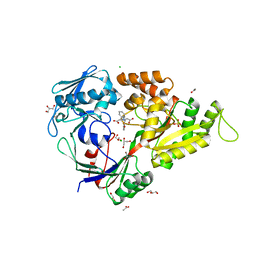 | | X-ray structure of a "NikA+Iron complex" hybrid, NikA/1 | | Descriptor: | 2-[2-[carboxymethyl(phenylmethyl)amino]ethyl-[(2-hydroxyphenyl)methyl]amino]ethanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cavazza, C, Bochot, C, Rousselot-Pailley, P, Carpentier, P, Cherrier, M.V, Martin, L, Marchi-Delapierre, C, Fontecilla-Camps, J.C, Menage, S. | | Deposit date: | 2010-05-05 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystallographic snapshots of the reaction of aromatic C-H with O(2) catalysed by a protein-bound iron complex
NAT.CHEM., 2, 2010
|
|
1JLF
 
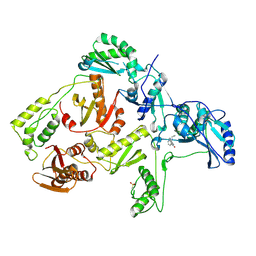 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLA
 
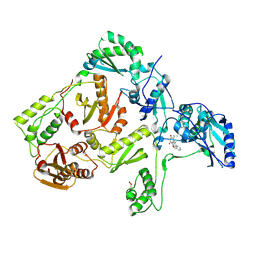 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLE
 
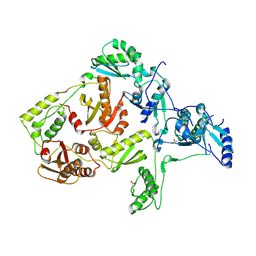 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | HIV-1 RT, A-CHAIN, B-CHAIN | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
4LEP
 
 | | Structural insights into substrate recognition in proton dependent oligopeptide transporters | | Descriptor: | N-[(1R)-1-phosphonoethyl]-L-alaninamide, Proton:oligopeptide symporter POT family, ZINC ION | | Authors: | Guettou, F, Quistgaard, E.M, Tresaugues, L, Moberg, P, Jegerschold, C, Zhu, L, Jong, A.J, Nordlund, P, Low, C. | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into substrate recognition in proton-dependent oligopeptide transporters.
Embo Rep., 14, 2013
|
|
6TJP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve - P212121 | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Fernandez, F.J, Machon, C, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Using a partial atomic model from medium-resolution cryo-EM to solve a large crystal structure.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1DZB
 
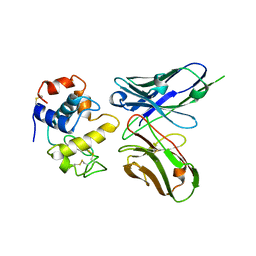 | | Crystal structure of phage library-derived single-chain Fv fragment 1F9 in complex with turkey egg-white lysozyme | | Descriptor: | SCFV FRAGMENT 1F9, TURKEY EGG-WHITE LYSOZYME C | | Authors: | Ay, J, Keitel, T, Kuettner, G, Wessner, H, Scholz, C, Hahn, M, Hoehne, W. | | Deposit date: | 2000-02-23 | | Release date: | 2000-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Phage Library-Derived Single-Chain Fv Fragment Complexed with Turkey Egg -White Lysozyme at 2.0 A Resolution
J.Mol.Biol., 301, 2000
|
|
1INI
 
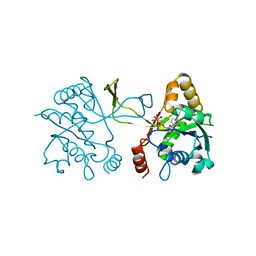 | | CRYSTAL STRUCTURE OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHETASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS, COMPLEXED WITH CDP-ME AND MG2+ | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHETASE, MAGNESIUM ION | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, A, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-05-14 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1INJ
 
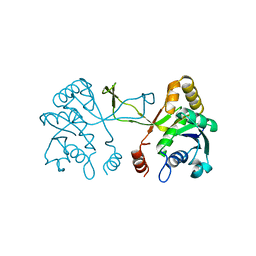 | | CRYSTAL STRUCTURE OF THE APO FORM OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHETASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHETASE, CALCIUM ION | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, A, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-05-14 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1I52
 
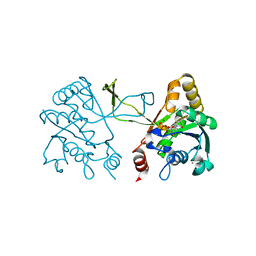 | | CRYSTAL STRUCTURE OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHASE, CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, M, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-02-23 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
7SR6
 
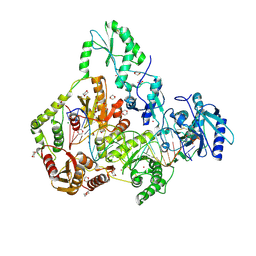 | | Human Endogenous Retrovirus (HERV-K) reverse transcriptase ternary complex with dsDNA template Primer and dNTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Baldwin, E.T, Nichols, C. | | Deposit date: | 2021-11-08 | | Release date: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Human endogenous retrovirus-K (HERV-K) reverse transcriptase (RT) structure and biochemistry reveals remarkable similarities to HIV-1 RT and opportunities for HERV-K-specific inhibition.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5I9K
 
 | | The structure of microsomal glutathione transferase 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTATHIONE, Microsomal glutathione S-transferase 1, ... | | Authors: | Kuang, Q, Purhonen, P, Jegerschold, C, Morgenstern, R, Hebert, H. | | Deposit date: | 2016-02-20 | | Release date: | 2017-07-12 | | Last modified: | 2017-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Dead-end complex, lipid interactions and catalytic mechanism of microsomal glutathione transferase 1, an electron crystallography and mutagenesis investigation.
Sci Rep, 7, 2017
|
|
5IA9
 
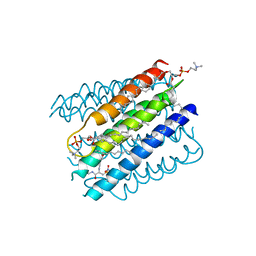 | | The structure of microsomal glutathione transferase 1 in complex with Meisenheimer complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Microsomal glutathione S-transferase 1, ... | | Authors: | Kuang, Q, Purhonen, P, Jegerschold, C, Morgenstern, R, Hebert, H. | | Deposit date: | 2016-02-21 | | Release date: | 2017-07-12 | | Last modified: | 2017-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Dead-end complex, lipid interactions and catalytic mechanism of microsomal glutathione transferase 1, an electron crystallography and mutagenesis investigation.
Sci Rep, 7, 2017
|
|
6QX5
 
 | | Crystal structure of T7 bacteriophage portal protein, 12mer, closed valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6R21
 
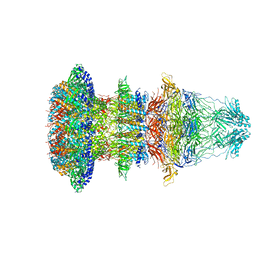 | | Cryo-EM structure of T7 bacteriophage fiberless tail complex | | Descriptor: | Portal protein, Tail tubular protein gp11, Tail tubular protein gp12 | | Authors: | Cuervo, A, Fabrega-Ferrer, M, Machon, C, Conesa, J.J, Perez-Ruiz, M, Coll, M, Carrascosa, J.L. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
1CFQ
 
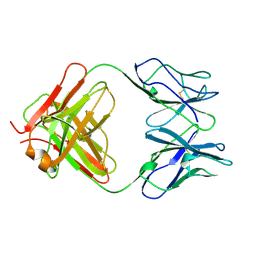 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 | | Descriptor: | PROTEIN (IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN)), PROTEIN (IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN)) | | Authors: | Keitel, T, Kramer, A, Wessner, H, Scholz, C, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 1999-03-19 | | Release date: | 1999-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of anti-p24 (HIV-1) monoclonal antibody cross-reactivity and polyspecificity.
Cell(Cambridge,Mass.), 91, 1997
|
|
1CFT
 
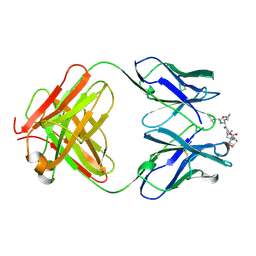 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH AN EPITOPE-UNRELATED D-PEPTIDE | | Descriptor: | PROTEIN (ANTIGEN BOUND PEPTIDE), PROTEIN (IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN)), PROTEIN (IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN)) | | Authors: | Keitel, T, Kramer, A, Wessner, H, Scholz, C, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 1999-03-19 | | Release date: | 1999-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of anti-p24 (HIV-1) monoclonal antibody cross-reactivity and polyspecificity.
Cell(Cambridge,Mass.), 91, 1997
|
|
1EP4
 
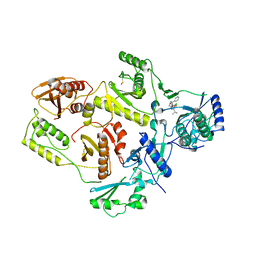 | | Crystal structure of HIV-1 reverse transcriptase in complex with S-1153 | | Descriptor: | 5-(3,5-DICHLOROPHENYL)THIO-4-ISOPROPYL-1-(PYRIDIN-4-YL-METHYL)-1H-IMIDAZOL-2-YL-METHYL CARBAMATE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Nichols, C, Bird, L.E, Fujiwara, T, Suginoto, H, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-03-27 | | Release date: | 2000-09-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding.
J.Biol.Chem., 275, 2000
|
|
3E3K
 
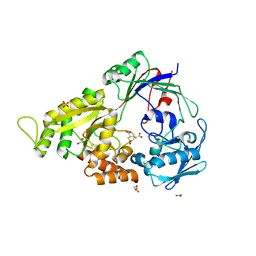 | | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA (butane-1,2,4-tricarboxylate without nickel form) | | Descriptor: | (2R)-butane-1,2,4-tricarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cherrier, M.V, Cavazza, C, Bochot, C, Lemaire, D, Fontecilla-Camps, J.C. | | Deposit date: | 2008-08-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA
Biochemistry, 47, 2008
|
|
3DP8
 
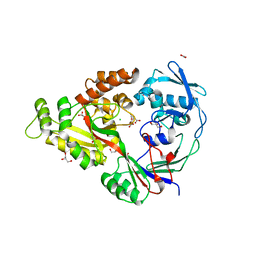 | | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA (nickel butane-1,2,4-tricarboxylate form) | | Descriptor: | (2R)-butane-1,2,4-tricarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cherrier, M.V, Cavazza, C, Bochot, C, Lemaire, D, Fontecilla-Camps, J.C. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA
Biochemistry, 47, 2008
|
|
4R02
 
 | | yCP in complex with BSc4999 (alpha-Keto Phenylamide) | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-{(2S,3S)-1-[(2,4-dimethylphenyl)amino]-2-hydroxy-5-methyl-1-oxohexan-3-yl}-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Voss, C, Scholz, C, Knorr, S, Beck, P, Stein, M, Zall, A, Kuckelkorn, U, Kloetzel, P.-M, Groll, M, Hamacher, K, Schmidt, B. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | alpha-Keto Phenylamides as P1'-Extended Proteasome Inhibitors.
Chemmedchem, 9, 2014
|
|
2XF4
 
 | | Crystal structure of Salmonella enterica serovar typhimurium YcbL | | Descriptor: | HYDROXYACYLGLUTATHIONE HYDROLASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Stamp, A, Owen, P, El Omari, K, Nichols, C, Lockyer, M, Lamb, H, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2010-05-20 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Salmonella Enterica Serovar Typhimurium Ycbl: An Unusual Type II Glyoxalase
Protein Sci., 19, 2010
|
|
2VAN
 
 | | Nucleotidyl Transfer Mechanism of Mismatched dNTP Incorporation by DNA Polymerase b by Structural and Kinetic Analyses | | Descriptor: | DNA POLYMERSE BETA | | Authors: | Chan, H, Chou, C, Tang, K, Niebuhr, M, Tung, C, Tsai, M. | | Deposit date: | 2007-09-03 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatched Dntp Incorporation by DNA Polymerase Beta Does not Proceed Via Globally Different Conformational Pathways.
Nucleic Acids Res., 36, 2008
|
|
3OBP
 
 | | Anaerobic complex of urate oxidase with uric acid | | Descriptor: | SODIUM ION, URIC ACID, Uricase | | Authors: | Gabison, L, Chopard, C, Colloc'h, N, El Hajji, M, Castro, B, Chiadmi, M, Prange, T. | | Deposit date: | 2010-08-08 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray, ESR, and quantum mechanics studies unravel a spin well in the cofactor-less urate oxidase.
Proteins, 79, 2011
|
|
8J46
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL | | Descriptor: | Olfactory receptor OR52c,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
