1C7R
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | 5-PHOSPHOARABINONIC ACID, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
1C7Q
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | N-BROMOACETYL-AMINOETHYL PHOSPHATE, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
8FUK
 
 | |
4M0W
 
 | | Crystal Structure of SARS-CoV papain-like protease C112S mutant in complex with ubiquitin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Replicase polyprotein 1a, ... | | Authors: | Chou, C.-Y, Chen, H.-Y, Lai, H.-Y, Cheng, S.-C, Chou, Y.-W. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5B7C
 
 | |
3LBC
 
 | |
3LCW
 
 | | L-KDO aldolase complexed with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-12 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
3LCF
 
 | | The D-sialic acid aldolase mutant V251I | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
3LCX
 
 | | L-KDO aldolase | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Huang, K.-F, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-12 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
3LCG
 
 | | The D-sialic acid aldolase mutant V251L | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
3LCI
 
 | | The D-sialic acid aldolase mutant V251W | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
3LCL
 
 | |
3LCH
 
 | | The D-sialic acid aldolase mutant V251R | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
5XU8
 
 | |
5XVE
 
 | |
3G8C
 
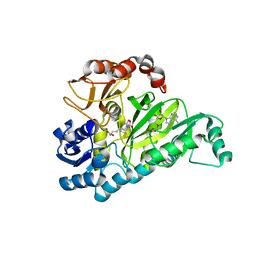 | | Crystal Structure of Biotin Carboxylase in Complex with Biotin, Bicarbonate, ADP and Mg Ion | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, BIOTIN, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
1UDV
 
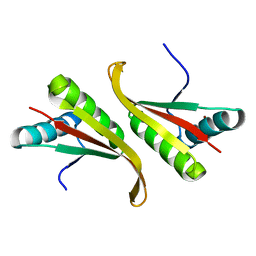 | | Crystal structure of the hyperthermophilic archaeal dna-binding protein Sso10b2 at 1.85 A | | Descriptor: | DNA binding protein SSO10b, ZINC ION | | Authors: | Chou, C.-C, Lin, T.-W, Chen, C.-Y, Wang, A.H.J. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 Angstroms
J.BACTERIOL., 185, 2003
|
|
3G8D
 
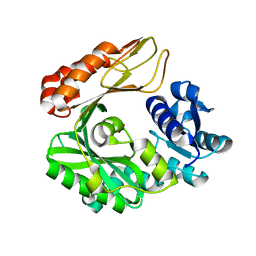 | | Crystal structure of the biotin carboxylase subunit, E296A mutant, of acetyl-COA carboxylase from Escherichia coli | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, MAGNESIUM ION, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
5C3N
 
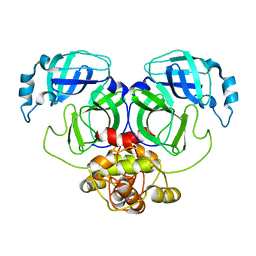 | |
3RV4
 
 | |
3RUP
 
 | |
3RV3
 
 | |
1UD0
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL 10-kDA SUBDOMAIN OF HSC70 | | Descriptor: | 70 kDa heat-shock-like protein, SODIUM ION | | Authors: | Chou, C.C, Forouhar, F, Yeh, Y.H, Wang, C, Hsiao, C.D. | | Deposit date: | 2003-04-24 | | Release date: | 2004-05-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the C-terminal 10-kDa subdomain of Hsc70
J.BIOL.CHEM., 278, 2003
|
|
2KVH
 
 | |
2KVF
 
 | |
