2XOA
 
 | |
4UJE
 
 | | Regulation of the mammalian elongation cycle by 40S subunit rolling: a eukaryotic-specific ribosome rearrangement | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN S10, ... | | Authors: | Budkevich, T.V, Giesebrecht, J, Behrmann, E, Loerke, J, Ramrath, D.J.F, Mielke, T, Ismer, J, Hildebrand, P, Tung, C.-S, Nierhaus, K.H, Sanbonmatsu, K.Y, Spahn, C.M.T. | | Deposit date: | 2014-04-05 | | Release date: | 2014-07-16 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement.
Cell(Cambridge,Mass.), 158, 2014
|
|
4CXG
 
 | | Regulation of the mammalian elongation cycle by 40S subunit rolling: a eukaryotic-specific ribosome rearrangement | | Descriptor: | 18S RRNA - H44, 18S RRNA - H5-H14, 18S RRNA - H8, ... | | Authors: | Budkevich, T.V, Giesebrecht, J, Behrmann, E, Loerke, J, Ramrath, D.J.F, Mielke, T, Ismer, J, Hildebrand, P, Tung, C.-S, Nierhaus, K.H, Sanbonmatsu, K.Y, Spahn, C.M.T. | | Deposit date: | 2014-04-07 | | Release date: | 2014-07-16 | | Last modified: | 2019-06-26 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement.
Cell(Cambridge,Mass.), 158, 2014
|
|
4CXH
 
 | | Regulation of the mammalian elongation cycle by 40S subunit rolling: a eukaryotic-specific ribosome rearrangement | | Descriptor: | 18S RRNA - H44, 18S RRNA - H5-H14, 18S RRNA - H8, ... | | Authors: | Budkevich, T.V, Giesebrecht, J, Behrmann, E, Loerke, J, Ramrath, D.J.F, Mielke, T, Ismer, J, Hildebrand, P, Tung, C.-S, Nierhaus, K.H, Sanbonmatsu, K.Y, Spahn, C.M.T. | | Deposit date: | 2014-04-07 | | Release date: | 2014-07-16 | | Last modified: | 2019-06-26 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement.
Cell(Cambridge,Mass.), 158, 2014
|
|
4DJC
 
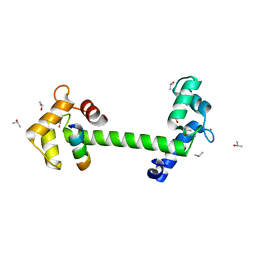 | | 1.35 A crystal structure of the NaV1.5 DIII-IV-Ca/CaM complex | | Descriptor: | CALCIUM ION, Calmodulin, ISOPROPYL ALCOHOL, ... | | Authors: | Sarhan, M.F, Tung, C.-C, Van Petegem, F, Ahern, C.A. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystallographic basis for calcium regulation of sodium channels.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5KJS
 
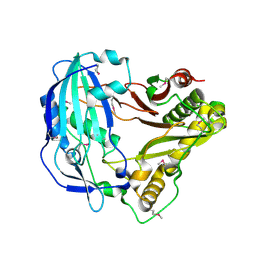 | | Crystal Structure of Arabidopsis thaliana HCT | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5K6P
 
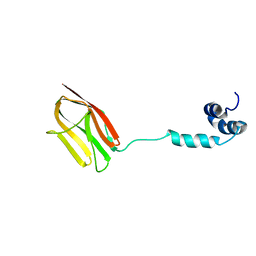 | | The NMR structure of the m domain tri-helix bundle and C2 of human cardiac Myosin Binding Protein C | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Michie, K.A, Kwan, A.H, Tung, C.S, Guss, J.M, Trewhella, J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-11-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure, 24, 2016
|
|
4L4H
 
 | |
5SV8
 
 | | Crystal Structure of the catalytic nucleophile and surface cysteine mutant of VvEG16 in complex with a xyloglucan oligosaccharide | | Descriptor: | alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, probable xyloglucan endotransglucosylase/hydrolase protein 19 | | Authors: | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | Deposit date: | 2016-08-05 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
5KJW
 
 | | Crystal structure of Coleus blumei HCT in complex with 3-hydroxyacetophenone | | Descriptor: | 1-(3-hydroxyphenyl)ethanone, Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJV
 
 | | Crystal structure of Coleus blumei HCT | | Descriptor: | Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJT
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroyl-CoA | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase, p-coumaroyl-CoA | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJU
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroylshikimate | | Descriptor: | (3~{R},4~{S},5~{R})-3-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-4,5-bis(oxidanyl)cyclohexene-1-carboxylic acid, Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5DZE
 
 | | Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with cellotetraose | | Descriptor: | beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, endo-glucanase | | Authors: | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | Deposit date: | 2015-09-25 | | Release date: | 2016-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
5DZF
 
 | | Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with a mixed-linkage glucan octasaccharide | | Descriptor: | SULFATE ION, beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | Deposit date: | 2015-09-25 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
5DZG
 
 | | Crystal Structure of the catalytic nucleophile mutant of VvEG16 in complex with a xyloglucan tetradecasaccharide | | Descriptor: | VvEG16, endo-glucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Tung, C.C, Van Petegem, F, Brumer, H. | | Deposit date: | 2015-09-25 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases.
Plant J., 89, 2017
|
|
6MUD
 
 | |
2VAN
 
 | | Nucleotidyl Transfer Mechanism of Mismatched dNTP Incorporation by DNA Polymerase b by Structural and Kinetic Analyses | | Descriptor: | DNA POLYMERSE BETA | | Authors: | Chan, H, Chou, C, Tang, K, Niebuhr, M, Tung, C, Tsai, M. | | Deposit date: | 2007-09-03 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatched Dntp Incorporation by DNA Polymerase Beta Does not Proceed Via Globally Different Conformational Pathways.
Nucleic Acids Res., 36, 2008
|
|
2VS6
 
 | | K173A, R174A, K177A-trichosanthin | | Descriptor: | RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Too, P.H, Ma, M.K, Mak, A.N, Tung, C.K, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C, Ng, A. | | Deposit date: | 2008-04-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
3JD5
 
 | | Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome | | Descriptor: | 28S ribosomal RNA, mitochondial, 28S ribosomal protein S10, ... | | Authors: | Kaushal, P.S, Sharma, M.R, Booth, T.M, Haque, E.M, Tung, C.S, Sanbonmatsu, K.Y, Spremulli, L.L, Agrawal, R.K. | | Deposit date: | 2016-04-08 | | Release date: | 2016-07-13 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2JJR
 
 | | V232K, N236D-trichosanthin | | Descriptor: | DI(HYDROXYETHYL)ETHER, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN, SULFATE ION, ... | | Authors: | Too, P.H, Ma, M.K, Mak, A.N, Tung, C.K, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C. | | Deposit date: | 2008-04-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
6B28
 
 | |
4I96
 
 | |
