6DRD
 
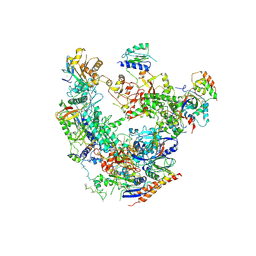 | | RNA Pol II(G) | | Descriptor: | DNA-directed RNA polymerase II subunit GRINL1A, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Yu, X, Jishage, M, Shi, Y, Ganesan, S, Sali, A, Chait, B.T, Asturias, F, Roeder, R.G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-12 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4FLG
 
 | | HIV-1 protease mutant I47V complexed with reaction intermediate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Yu, X, Shen, C.H, Weber, I.T. | | Deposit date: | 2012-06-14 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Capturing the Reaction Pathway in Near-Atomic-Resolution Crystal Structures of HIV-1 Protease.
Biochemistry, 51, 2012
|
|
8E8O
 
 | | Cryo-EM structure of human ME3 in the presence of citrate | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8E76
 
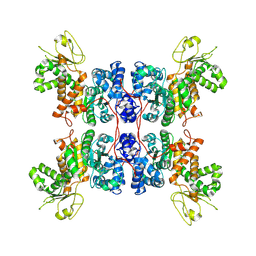 | | Cryo-EM structure of Apo form ME3 | | Descriptor: | NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8E78
 
 | | Cryo-EM structure of human ME3 in the presence of citrate | | Descriptor: | NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
7S78
 
 | |
5TX1
 
 | | Cryo-Electron microscopy structure of species-D human adenovirus 26 | | Descriptor: | Fiber, Hexon protein, PIIIa, ... | | Authors: | Reddy, V, Yu, X, Veesler, D. | | Deposit date: | 2016-11-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses.
Sci Adv, 3, 2017
|
|
3SIS
 
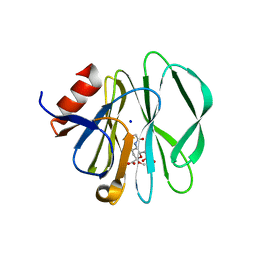 | |
3SIT
 
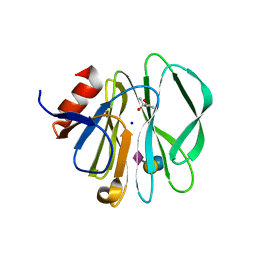 | |
5BWI
 
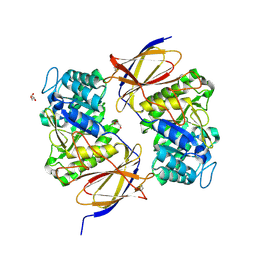 | |
3TJZ
 
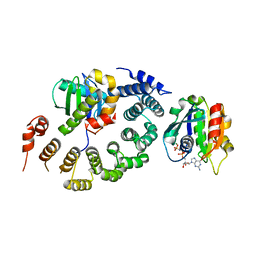 | | Crystal Structure of Arf1 Bound to the gamma/zeta-COP Core Complex | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit gamma, Coatomer subunit zeta-1, ... | | Authors: | Goldberg, J, Yu, X, Breitman, M. | | Deposit date: | 2011-08-25 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structure-based mechanism for arf1-dependent recruitment of coatomer to membranes.
Cell(Cambridge,Mass.), 148, 2012
|
|
2QCJ
 
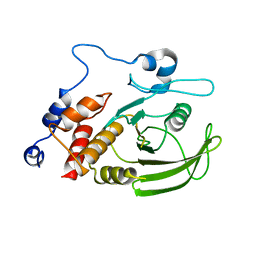 | | Native Structure of Lyp | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Sun, J.P, Yu, X, Zhang, Z.Y. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Lyp and its complex with a selective inhibitor
To be Published
|
|
8F0G
 
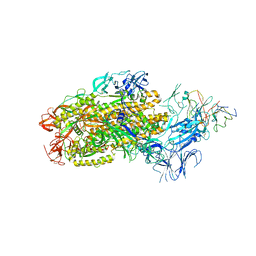 | | Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody Fab 1C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 1C3 Fab Heavy Chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8F0H
 
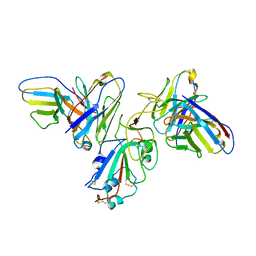 | | Structure of SARS-CoV-2 spike with antibody Fabs 2A10 and 1H2 (Local refinement of the RBD and Fabs 1H2 and 2A10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 1H2 heavy chain, Antibody Fab 1H2 light chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
7WGR
 
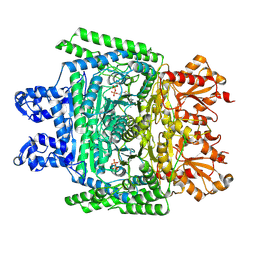 | | Cryo-electron microscopic structure of the 2-oxoglutarate dehydrogenase (E1) component of the human alpha-ketoglutarate (2-oxoglutarate) dehydrogenase complex | | Descriptor: | 2-oxoglutarate dehydrogenase, mitochondrial, CALCIUM ION, ... | | Authors: | Yu, X, Yang, W, Zhong, Y.H, Ma, X.M, Gao, Y.Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis for the activity and regulation of human alpha-ketoglutarate dehydrogenase revealed by Cryo-EM
Biochem.Biophys.Res.Commun., 602, 2022
|
|
2G7M
 
 | | Crystal structure of B. fragilis N-succinylornithine transcarbamylase P90E mutant complexed with carbamoyl phosphate and N-acetylnorvaline | | Descriptor: | N-ACETYL-L-NORVALINE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, SULFATE ION, ... | | Authors: | Shi, D, Yu, X, Roth, L, Morizono, H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2006-02-28 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
5KUA
 
 | | Cryo-EM reconstruction of Neisseria meningitidis Type IV pilus | | Descriptor: | pilin | | Authors: | Kolappan, S, Coureuil, M, Yu, X, Nassif, X, Craig, L, Egelman, E.H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-12 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Neisseria meningitidis Type IV pilus.
Nat Commun, 7, 2016
|
|
1OS5
 
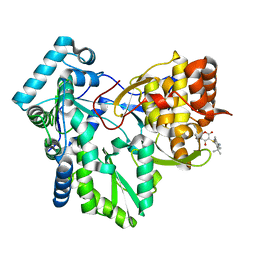 | | Crystal structure of HCV NS5B RNA polymerase complexed with a novel non-competitive inhibitor. | | Descriptor: | 3-(4-AMINO-2-TERT-BUTYL-5-METHYL-PHENYLSULFANYL)-6-CYCLOPENTYL-4-HYDROXY-6-[2-(4-HYDROXY-PHENYL)-ETHYL]-5,6-DIHYDRO-PYRAN-2-ONE, Hepatitis C virus NS5B RNA polymerase | | Authors: | Love, R.A, Parge, H.E, Yu, X, Hickey, M.J, Diehl, W, Gao, J, Wriggers, H, Ekker, A, Wang, L, Thomson, J.A, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2003-03-18 | | Release date: | 2004-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme.
J.Virol., 77, 2003
|
|
7PLN
 
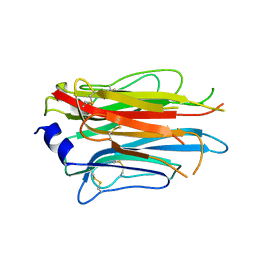 | | Structure of the APCbeta domain of Plasmodium vivax perforin-like protein 1 | | Descriptor: | Sporozoite micronemal protein essential for cell traversal, putative | | Authors: | Williams, S.I, Ni, T, Yu, X, Gilbert, R.J.C. | | Deposit date: | 2021-08-31 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural, Functional and Computational Studies of Membrane Recognition by Plasmodium Perforin-Like Proteins 1 and 2
J.Mol.Biol., 434, 2022
|
|
6RYO
 
 | | Bacterial membrane enzyme structure by the in meso method at 1.9 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, GLYCEROL, ... | | Authors: | Huang, C.Y, Olatunji, S, Bailey, J, Yu, X, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | Structures of lipoprotein signal peptidase II from Staphylococcus aureus complexed with antibiotics globomycin and myxovirescin.
Nat Commun, 11, 2020
|
|
1WT9
 
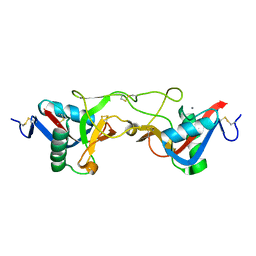 | | crystal structure of Aa-X-bp-I, a snake venom protein with the activity of binding to coagulation factor X from Agkistrodon acutus | | Descriptor: | CALCIUM ION, agkisacutacin A chain, anticoagulant protein-B | | Authors: | Zhu, Z, Liu, S, Mo, X, Yu, X, Liang, Z, Zang, J, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2004-11-18 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterizations and Crystal structures of two snake venom proteins with the activity of binding coagulation factor X from Agkistrodon acutus
To be Published
|
|
6SB3
 
 | |
6SB4
 
 | | Crystal structure of murine perforin-2 P2 domain crystal form 2 | | Descriptor: | Macrophage-expressed gene 1 protein | | Authors: | Ni, T, Yu, X, Ginger, L, Gilbert, R.J.C. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity.
Sci Adv, 6, 2020
|
|
7BR8
 
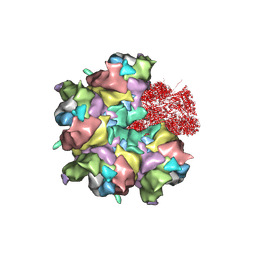 | | Epstein-Barr virus, C5 penton vertex, CATC absent. | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
6SB5
 
 | |
