1DF0
 
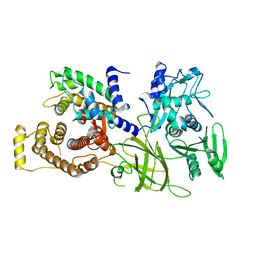 | | Crystal structure of M-Calpain | | Descriptor: | CALPAIN, M-CALPAIN | | Authors: | Hosfield, C.M, Elce, J.S, Davies, P.L, Jia, Z. | | Deposit date: | 1999-11-16 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation.
EMBO J., 18, 1999
|
|
1DKL
 
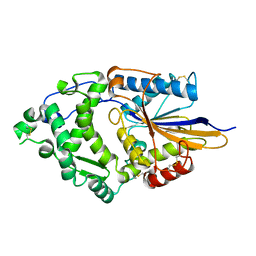 | |
1DKO
 
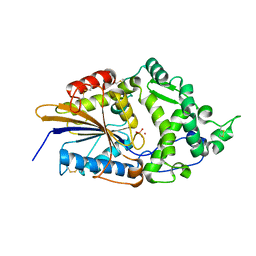 | | CRYSTAL STRUCTURE OF TUNGSTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH TUNGSTATE BOUND AT THE ACTIVE SITE AND WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE, TUNGSTATE(VI)ION | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKM
 
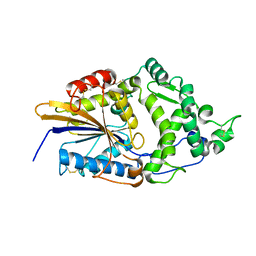 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKQ
 
 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX ESCHERICHIA COLI PHYTASE AT PH 5.0. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKP
 
 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKN
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 5.0 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
2MSI
 
 | |
6XCL
 
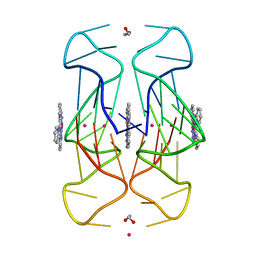 | | Crystal Structure of human telomeric DNA G-quadruplex in complex with a novel platinum(II) complex. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), POTASSIUM ION, ... | | Authors: | Miron, C.E, van Staalduinen, L.M, Jia, Z, Petitjean, A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Going Platinum to the Tune of a Remarkable Guanine Quadruplex Binder: Solution- and Solid-State Investigations.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2JIA
 
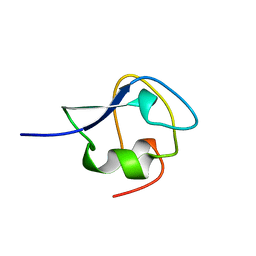 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 K61I | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1NP8
 
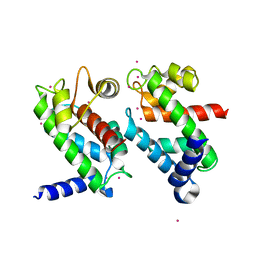 | | 18-k C-terminally trunucated small subunit of calpain | | Descriptor: | CADMIUM ION, Calcium-dependent protease, small subunit | | Authors: | Leinala, E.K, Arthur, J.S, Grochulski, P, Davies, P.L, Elce, J.S, Jia, Z. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A second binding site revealed by C-terminal truncation of calpain small subunit, a penta-EF-hand protein
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
1NT4
 
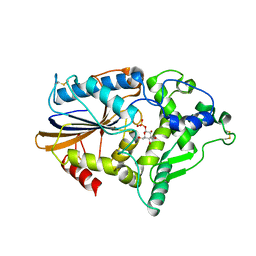 | | Crystal structure of Escherichia coli periplasmic glucose-1-phosphatase H18A mutant complexed with glucose-1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-glucopyranose, Glucose-1-phosphatase | | Authors: | Lee, D.C, Cottrill, M.A, Forsberg, C.W, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-01-28 | | Release date: | 2004-01-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights revealed by the crystal structures of Escherichia coli glucose-1-phosphatase.
J.Biol.Chem., 278, 2003
|
|
1QXP
 
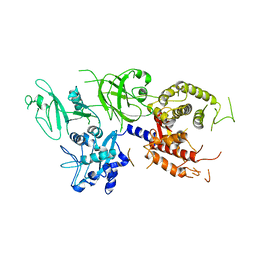 | |
2ASI
 
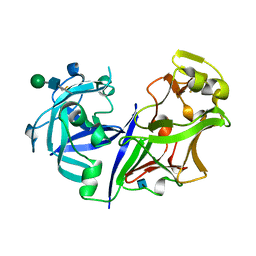 | | ASPARTIC PROTEINASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARTIC PROTEINASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Jia, Z, Vandonselaar, M, Kepliakov, P.S.A, Quail, J.W. | | Deposit date: | 1995-12-09 | | Release date: | 1996-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the aspartic proteinase from Rhizomucor miehei at 2.15 A resolution.
J.Mol.Biol., 268, 1997
|
|
3HOK
 
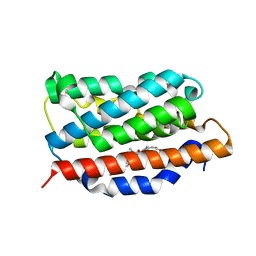 | | X-ray Crystal Structure of Human Heme Oxygenase-1 with (2R, 4S)-2-[2-(4-Chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-4[((5-trifluoromethylpyridin-2-yl)thio)methyl]-1,3-dioxolane: A Novel, Inducible Binding Mode | | Descriptor: | 2-({[(2R,4S)-2-[2-(4-chlorophenyl)ethyl]-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methyl}sulfanyl)-5-(trifluoromethyl)pyridine, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rahman, M.N, Jia, Z. | | Deposit date: | 2009-06-02 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystal structure of human heme oxygenase-1 with (2R,4S)-2-[2-(4-chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-4[((5-trifluoromethylpyridin-2-yl)thio)methyl]-1,3-dioxolane: a novel, inducible binding mode.
J.Med.Chem., 52, 2009
|
|
3I3L
 
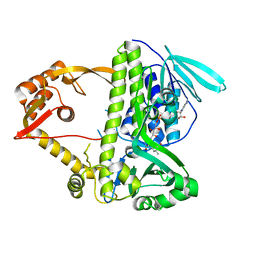 | | Crystal structure of CmlS, a flavin-dependent halogenase | | Descriptor: | Alkylhalidase CmlS, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Podzelinska, K, Soares, A, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-06-30 | | Release date: | 2010-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chloramphenicol Biosynthesis: The Structure of CmlS, a Flavin-Dependent Halogenase Showing a Covalent Flavin-Aspartate Bond
J.Mol.Biol., 397, 2010
|
|
3KCP
 
 | | Crystal structure of interacting Clostridium thermocellum multimodular components | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cellulosomal-scaffolding protein A, ... | | Authors: | Adams, J.J, Currie, M.A, Bayer, E.A, Jia, Z, Smith, S.P. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into Higher-Order Organization of the Cellulosome Revealed by a Dissect-and-Build Approach: Crystal Structure of Interacting Clostridium thermocellum Multimodular Components
J.Mol.Biol., 396, 2010
|
|
3KMH
 
 | |
3K4F
 
 | | X-Ray Crystal Structure of Human Heme Oxygenase-1 in Complex with 4-Phenyl-1-(1H-1,2,4-triazol-1-yl)-2-butanone | | Descriptor: | 4-phenyl-1-(1H-1,2,4-triazol-1-yl)butan-2-one, HEXANE-1,6-DIOL, Heme oxygenase 1, ... | | Authors: | Rahman, M.N, Jia, Z. | | Deposit date: | 2009-10-05 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Heme Oxygenase Inhibition by 2-Oxy-substituted 1-Azolyl-4-phenylbutanes: Effect of Variation of the Azole Moiety. X-Ray Crystal Structure of Human Heme Oxygenase-1 in Complex with 4-Phenyl-1-(1H-1,2,4-triazol-1-yl)-2-butanone.
Chem.Biol.Drug Des., 75, 2010
|
|
2OVI
 
 | |
2R1A
 
 | |
2R19
 
 | |
6UTS
 
 | |
6UTU
 
 | | Crystal structure of minor pseudopilin ternary complex of XcpVWX from the Type 2 secretion system of Pseudomonas aeruginosa in the P3 space group | | Descriptor: | CALCIUM ION, Type II secretion system protein I, Type II secretion system protein J, ... | | Authors: | Zhang, Y, Wang, S, Jia, Z. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | In Situ Proteolysis Condition-Induced Crystallization of the XcpVWX Complex in Different Lattices.
Int J Mol Sci, 21, 2020
|
|
1EWW
 
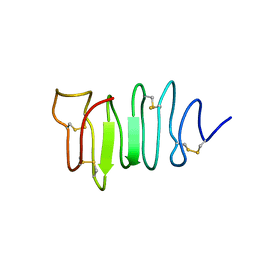 | | SOLUTION STRUCTURE OF SPRUCE BUDWORM ANTIFREEZE PROTEIN AT 30 DEGREES CELSIUS | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Graether, S.P, Kuiper, M.J, Gagne, S.M, Walker, V.K, Jia, Z, Sykes, B.D, Davies, P.L. | | Deposit date: | 2000-04-27 | | Release date: | 2000-07-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Beta-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect.
Nature, 406, 2000
|
|
