5NML
 
 | | Nb36 Ser85Cys with Hg bound | | Descriptor: | 1,2-ETHANEDIOL, MERCURY (II) ION, Nanobody Nb36 Ser85Cys | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NLU
 
 | | Structure of Nb36 crystal form 1 | | Descriptor: | SULFATE ION, single domain llama antibody Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NM0
 
 | | Nb36 Ser85Cys with Hg, crystal form 1 | | Descriptor: | MERCURY (II) ION, Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1N0V
 
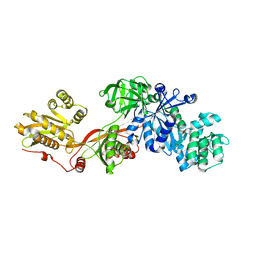 | | Crystal structure of elongation factor 2 | | Descriptor: | Elongation factor 2 | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat.Struct.Biol., 10, 2003
|
|
1N0U
 
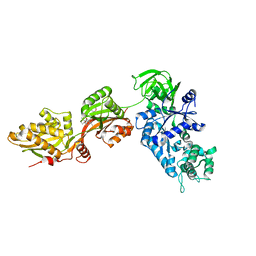 | | Crystal structure of yeast elongation factor 2 in complex with sordarin | | Descriptor: | Elongation factor 2, [1R-(1.ALPHA.,3A.BETA.,4.BETA.,4A.BETA.,7.BETA.,7A.ALPHA.,8A.BETA.)]8A-[(6-DEOXY-4-O-METHYL-BETA-D-ALTROPYRANOSYLOXY)METHYL]-4-FORMYL-4,4A,5,6,7,7A,8,8A-OCTAHYDRO-7-METHYL-3-(1-METHYLETHYL)-1,4-METHANO-S-INDACENE-3A(1H)-CARBOXYLIC ACID | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat. Struct. Biol., 10, 2003
|
|
7P2D
 
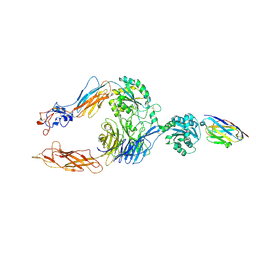 | | Structure of alphaMbeta2/Cd11bCD18 headpiece in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2021-07-05 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the function-modulating effects of nanobody binding to the integrin receptor alpha M beta 2.
J.Biol.Chem., 298, 2022
|
|
6SEJ
 
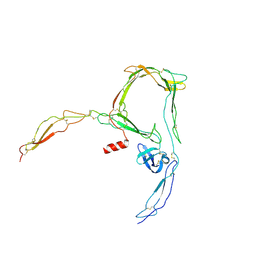 | | Structure of a functional monomeric properdin lacking TSR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Properdin, alpha-D-mannopyranose, ... | | Authors: | Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System.
Front Immunol, 10, 2019
|
|
6RV6
 
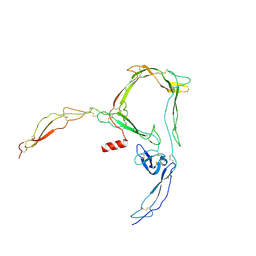 | | Structure of properdin lacking TSR3 based on anomalous data | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Properdin, alpha-D-mannopyranose, ... | | Authors: | Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2019-05-31 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.507 Å) | | Cite: | Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System.
Front Immunol, 10, 2019
|
|
6RUR
 
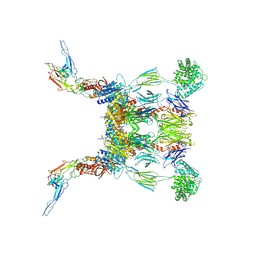 | | Structure of the SCIN stabilized C3bBb convertase bound to properdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, ... | | Authors: | Pedersen, D.V, Gadeberg, T.A.F, Andersen, G.R. | | Deposit date: | 2019-05-29 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System.
Front Immunol, 10, 2019
|
|
3KX2
 
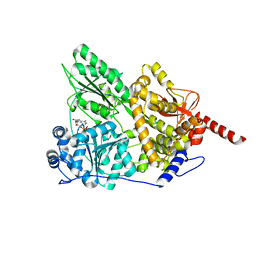 | |
3MMP
 
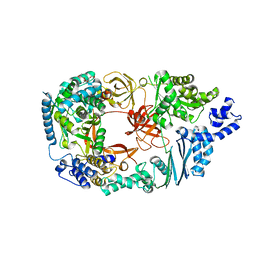 | | Structure of the Qb replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Elongation factor Tu 2, Elongation factor Ts, ... | | Authors: | Kidmose, R.T, Vasiliev, N.N, Chetverin, A.B, Knudsen, C.R, Andersen, G.R. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KLS
 
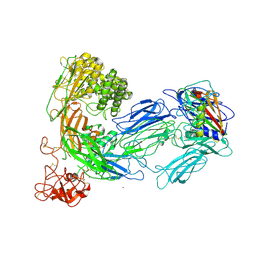 | | Structure of complement C5 in complex with SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KM9
 
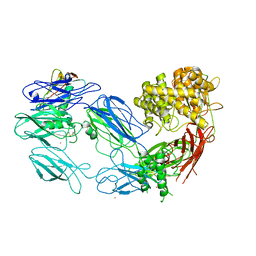 | | Structure of complement C5 in complex with the C-terminal beta-grasp domain of SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4X7S
 
 | |
4XW2
 
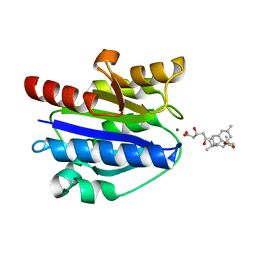 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | Descriptor: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | Authors: | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | Deposit date: | 2015-01-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
4X7T
 
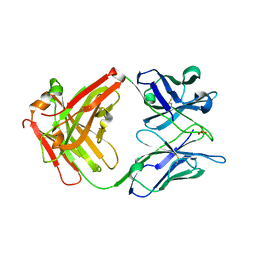 | |
4YBH
 
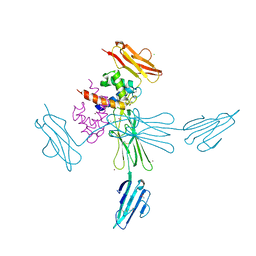 | |
8COH
 
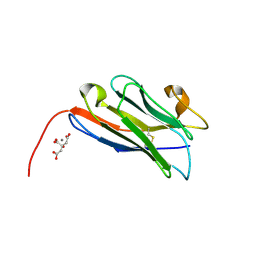 | | Structure of the complement C5 specific nanobody TPP-3444 | | Descriptor: | CITRIC ACID, MANGANESE (II) ION, Nanobody TPP-3444 | | Authors: | Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of the bispecific VHH antibody gefurulimab (ALXN1720) targeting complement component 5, and designed for low volume subcutaneous administration.
Mol.Immunol., 165, 2023
|
|
2P8Z
 
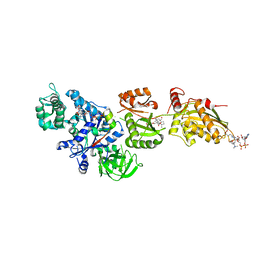 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2P8W
 
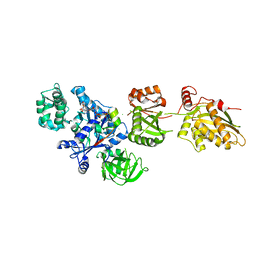 | | Fitted structure of eEF2 in the 80S:eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | Elongation factor 2, Elongation factor Tu-B, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
4HWJ
 
 | |
6YSQ
 
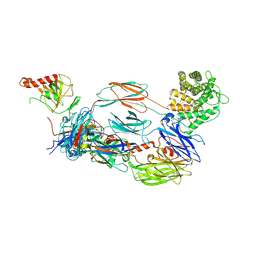 | | The hC4Nb8 complement inhibitory nanobody in complex with C4b | | Descriptor: | Complement C4 beta chain, Complement C4 gamma chain, Complement C4-A alpha chain, ... | | Authors: | Zarantonello, A, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | An Ultrahigh-Affinity Complement C4b-Specific Nanobody Inhibits In Vivo Assembly of the Classical Pathway Proconvertase.
J Immunol., 205, 2020
|
|
2P8X
 
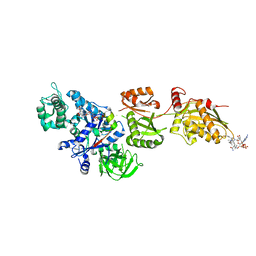 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
4HW5
 
 | |
2P8Y
 
 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
