8ZAX
 
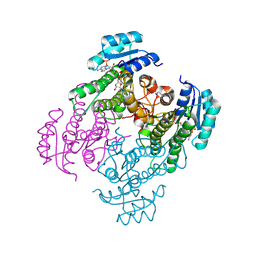 | | Crystal structure of a short-chain dehydrogenase from Lactobacillus fermentum with NADPH | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SDR family oxidoreductase | | Authors: | Wang, J.J, Cong, L, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-04-25 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir
Adv.Synth.Catal., 2024
|
|
6DJC
 
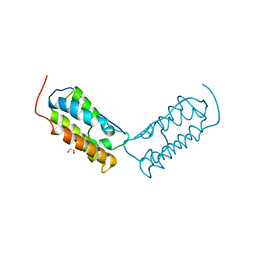 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8YAV
 
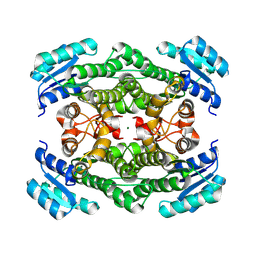 | | Crystal structure of glucose 1-dehydrogenase from Limosilactobacillus fermentum | | Descriptor: | MAGNESIUM ION, SDR family oxidoreductase | | Authors: | Cong, L, Wang, J.J, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-10 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir.
Adv.Synth.Catal., n/a, 2024
|
|
8YAI
 
 | | Crystal structure of glucose 1-dehydrogenase mutant1 from Limosilactobacillus fermentum | | Descriptor: | SDR family oxidoreductase | | Authors: | Cong, L, Wang, J.J, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir
Adv.Synth.Catal., 2024
|
|
8YAU
 
 | |
6LJ1
 
 | |
6LJ2
 
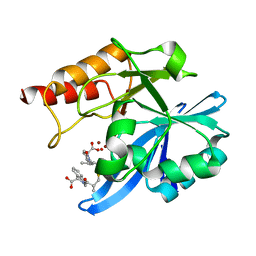 | | Crystal structure of NDM-1 in complex with heterodimer of D-captopril derivative wss02127 stereoisomer | | Descriptor: | (1R)-2-[(2S)-2-methyl-3-sulfanyl-propanoyl]-3,4-dihydro-1H-isoquinoline-1-carboxylic acid, (1S)-2-[(2S)-2-methyl-3-sulfanyl-propanoyl]-3,4-dihydro-1H-isoquinoline-1-carboxylic acid, HYDROXIDE ION, ... | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJ5
 
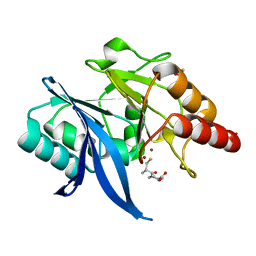 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss04145 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJ0
 
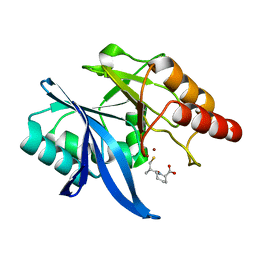 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss02122 | | Descriptor: | (2R)-1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidine-2-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJ6
 
 | |
6LJ4
 
 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss04146 | | Descriptor: | (3S)-1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidine-3-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJ8
 
 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss04134 | | Descriptor: | 1,2-ETHANEDIOL, 2-[1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidin-4-yl]ethanoic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LIP
 
 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss0218 | | Descriptor: | (2R)-1-[3-sulfanyl-2-(sulfanylmethyl)propanoyl]pyrrolidine-2-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LIZ
 
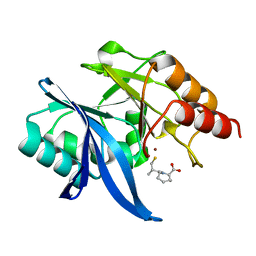 | |
6LJ7
 
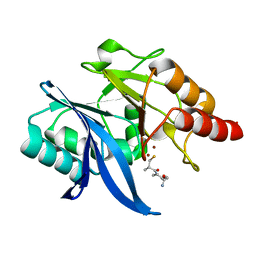 | |
4V68
 
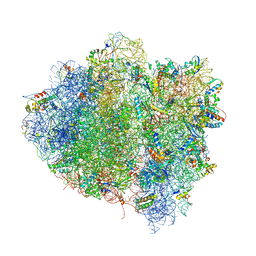 | | T. thermophilus 70S ribosome in complex with mRNA, tRNAs and EF-Tu.GDP.kirromycin ternary complex, fitted to a 6.4 A Cryo-EM map. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schuette, J.-C, Spahn, C.M.T. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | GTPase activation of elongation factor EF-Tu by the ribosome during decoding
Embo J., 28, 2009
|
|
4N6Z
 
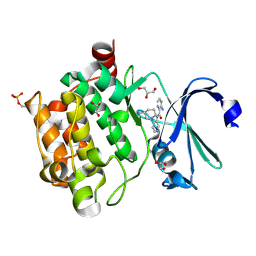 | | Pim1 Complexed with a pyridylcarboxamide | | Descriptor: | 3-amino-N-{4-[(3S)-3-aminopiperidin-1-yl]pyridin-3-yl}pyrazine-2-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4W9N
 
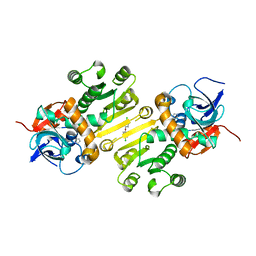 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase complexed with triclosan | | Descriptor: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, IMIDAZOLE, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
4N6Y
 
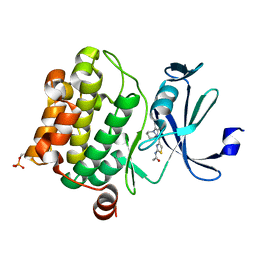 | | Pim1 Complexed with a phenylcarboxamide | | Descriptor: | 2-(acetylamino)-N-[2-(piperidin-1-yl)phenyl]-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
7MBO
 
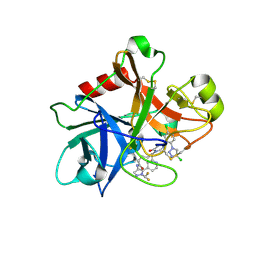 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.924 Å) | | Cite: | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
4N70
 
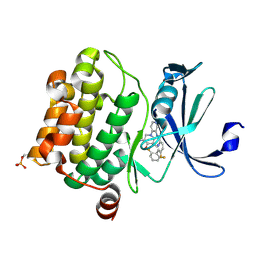 | | Pim1 Complexed with a pyridylcarboxamide | | Descriptor: | N-{4-[(3R,4R,5S)-3-amino-4-hydroxy-5-methylpiperidin-1-yl]pyridin-3-yl}-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
7RIT
 
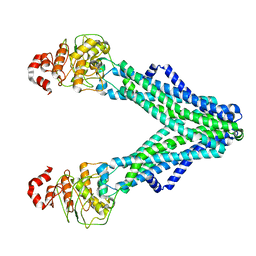 | | Drug-free A. baumannii MsbA | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-07-20 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
8GQ6
 
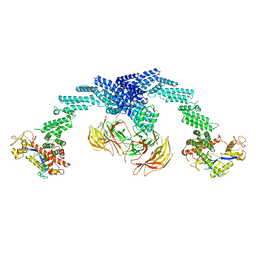 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.51 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
