3IIY
 
 | | Crystal structure of Eed in complex with a trimethylated histone H1K26 peptide | | Descriptor: | Histone H1K26 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IIW
 
 | | Crystal structure of Eed in complex with a trimethylated histone H3K27 peptide | | Descriptor: | Histone H3 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJ0
 
 | | Crystal structure of Eed in complex with a trimethylated histone H3K9 peptide | | Descriptor: | Histone H3K9 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJC
 
 | | Crystal structure of Eed in complex with NDSB-195 | | Descriptor: | ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJ1
 
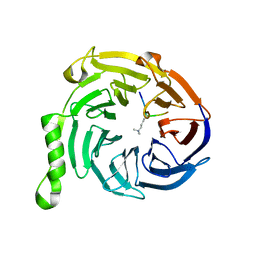 | | Crystal structure of Eed in complex with a trimethylated histone H4K20 peptide | | Descriptor: | Histone H4K20 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
5ODF
 
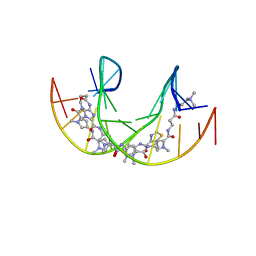 | |
5OE1
 
 | | Im polyamide in complex with 5'CGATGTACATCG3'- hairpin polyamides studies | | Descriptor: | 1-methylimidazole-2-carboxylic acid, 3-(3-azaniumylpropanoylamino)propyl-dimethyl-azanium, 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, ... | | Authors: | Padroni, G, Parkinson, J, Burley, G.A. | | Deposit date: | 2017-07-07 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides.
Nucleic Acids Res., 46, 2018
|
|
7LBC
 
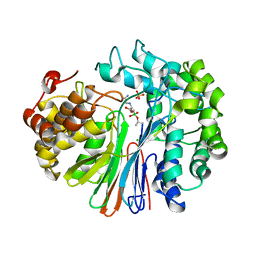 | | Structure of human GGT1 in complex with Lnt2-65 compound | | Descriptor: | (2R)-2-[(2-aminoethyl)amino]-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-07 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
7LD9
 
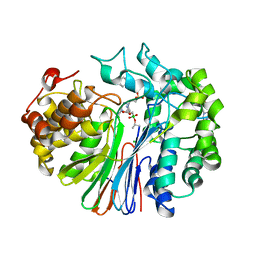 | | Structure of human GGT1 in complex with ABBA | | Descriptor: | (2R)-2-amino-4-boronobutanoic acid, (2S)-2-amino-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-12 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
5OFT
 
 | |
3K48
 
 | | Crystal structure of APRIL bound to a peptide | | Descriptor: | Tumor necrosis factor ligand superfamily member 13, peptide | | Authors: | Hymowitz, S.G. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Multiple novel classes of APRIL-specific receptor-blocking peptides isolated by phage display.
J.Mol.Biol., 396, 2010
|
|
4N23
 
 | | Crystal structure of the GP2 Core Domain from the California Academy of Science Virus, monoclinic symmetry | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GP2 Ectodomain | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Dai, Z, Toro, R, Lai, J.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species.
J.Mol.Biol., 426, 2014
|
|
2PR3
 
 | | Factor XA inhibitor | | Descriptor: | (2R,4R)-N~1~-(4-CHLOROPHENYL)-N~2~-[3-FLUORO-2'-(METHYLSULFONYL)BIPHENYL-4-YL]-4-METHOXYPYRROLIDINE-1,2-DICARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Zhang, E, Kohrt, J.T, Bigge, C.F, Finzel, B.C. | | Deposit date: | 2007-05-03 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based drug design of pyrrolidine-1, 2-dicarboxamides as a novel series of orally bioavailable factor Xa inhibitors
Chem.Biol.Drug Des., 69, 2007
|
|
2Q2B
 
 | |
3ZZP
 
 | | Circular permutant of ribosomal protein S6, lacking edge strand beta- 2 of wild-type S6. | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Saraboji, K, Haglund, E, Lindberg, M.O, Oliveberg, M, Logan, D.T. | | Deposit date: | 2011-09-02 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Trimming Down a Protein Structure to its Bare Foldons: Spatial Organization of the Cooperative Unit.
J.Biol.Chem., 287, 2012
|
|
2ANR
 
 | | Crystal structure (II) of Nova-1 KH1/KH2 domain tandem with 25nt RNA hairpin | | Descriptor: | 5'-R(*CP*(5BU)P*CP*GP*CP*GP*GP*AP*UP*CP*AP*GP*UP*CP*AP*CP*CP*CP*AP*AP*GP*CP*GP*AP*G)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Malinina, L, Teplova, M, Musunuru, K, Teplov, A, Darnell, J.C, Burley, S.K, Darnell, R.B, Patel, D.J. | | Deposit date: | 2005-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1.
Structure, 19, 2011
|
|
8K37
 
 | | Structure of the bacteriophage lambda neck | | Descriptor: | Head-tail connector protein FII, Tail tube protein, Tail tube terminator protein | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K36
 
 | |
8K35
 
 | | Structure of the bacteriophage lambda tail tip complex | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K38
 
 | | The structure of bacteriophage lambda portal-adaptor | | Descriptor: | Head completion protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K39
 
 | | Structure of the bacteriophage lambda portal vertex | | Descriptor: | Major capsid protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8QUS
 
 | |
8QU7
 
 | |
5WG5
 
 | | Human GRK2 in complex with Gbetagamma subunits and CCG224061 | | Descriptor: | 5-{[(3S,4R)-4-(4-fluorophenyl)piperidin-3-yl]methoxy}-2H-indazole, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Bouley, R, Tesmer, J.J.G. | | Deposit date: | 2017-07-13 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Determinants Influencing the Potency and Selectivity of Indazole-Paroxetine Hybrid G Protein-Coupled Receptor Kinase 2 Inhibitors.
Mol. Pharmacol., 92, 2017
|
|
5WG4
 
 | | Human GRK2 in complex with Gbetagamma subunits and CCG257284 | | Descriptor: | 2-fluoro-5-[(3S,4R)-3-{[(1H-indazol-5-yl)oxy]methyl}piperidin-4-yl]-N-[(pyridin-2-yl)methyl]benzamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Bouley, R, Tesmer, J.J.G. | | Deposit date: | 2017-07-13 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Determinants Influencing the Potency and Selectivity of Indazole-Paroxetine Hybrid G Protein-Coupled Receptor Kinase 2 Inhibitors.
Mol. Pharmacol., 92, 2017
|
|
