8VW4
 
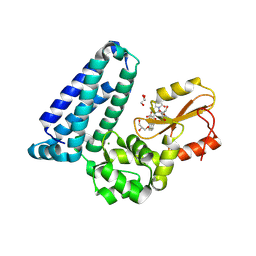 | | Crystal structure of Cbl-b TKB bound to compound 26 | | Descriptor: | (7-methoxy-2-{2-[(1S,3S,4S)-3-(3-methoxy-2-methyl-5-nitrophenyl)-1-methyl-5-oxo-1,5-dihydroimidazo[1,5-a]pyridin-2(3H)-yl]-2-oxoethoxy}quinolin-8-yl)acetic acid, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Yu, C, Murray, J, Hsu, P.L. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
5MF0
 
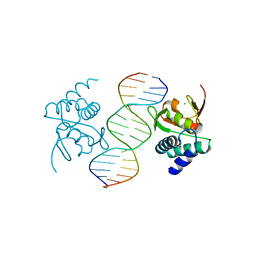 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5NM9
 
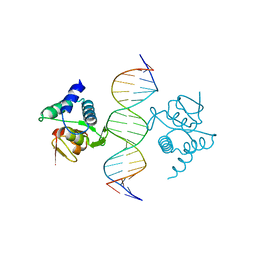 | | Crystal structure of the placozoa Trichoplax adhaerens Smad4-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*AP*TP*GP*CP*GP*GP*GP*CP*GP*CP*GP*CP*CP*CP*GP*CP*AP*T)-3'), Mothers against decapentaplegic homolog, ZINC ION | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5ODG
 
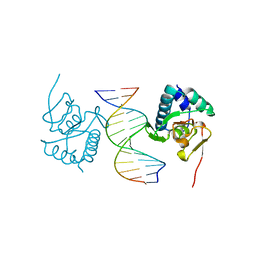 | | Crystal structure of Smad3-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 3, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | Authors: | Wang, K, Wang, X, Pan, L. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
5GV2
 
 | |
6LQZ
 
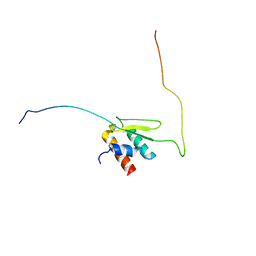 | | Solution structure of Taf14ET-Sth1EBMC | | Descriptor: | Nuclear protein STH1/NPS1, Transcription initiation factor TFIID subunit 14 | | Authors: | Wu, B, Chen, G, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation.
Nat Commun, 11, 2020
|
|
4A2B
 
 | |
4A2A
 
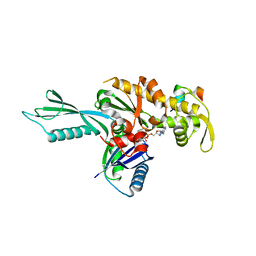 | | Thermotoga maritima FtsA:FtsZ(336-351) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN FTSA, PUTATIVE, ... | | Authors: | Szwedziak, P, Lowe, J. | | Deposit date: | 2011-09-23 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ftsa Forms Actin-Like Protofilaments
Embo J., 31, 2012
|
|
4B46
 
 | |
4B45
 
 | |
5ISY
 
 | | Crystal structure of Nudix family protein with NAD | | Descriptor: | NADH pyrophosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-15 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural basis of prokaryotic NAD-RNA decapping by NudC
Cell Res., 26, 2016
|
|
3ZID
 
 | |
6WO1
 
 | |
7V61
 
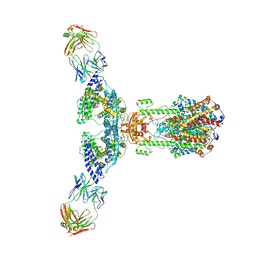 | | ACE2 -Targeting Monoclonal Antibody as Potent and Broad-Spectrum Coronavirus Blocker | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3E8, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker.
Signal Transduct Target Ther, 6, 2021
|
|
5H0N
 
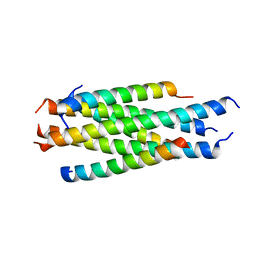 | |
7F9W
 
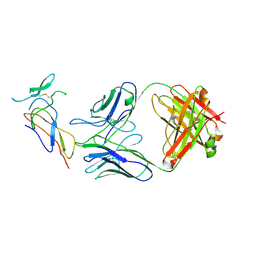 | | CD25 in complex with Fab | | Descriptor: | Heavy chain of Fab, Interleukin-2 receptor subunit alpha, Light chain of Fab | | Authors: | Liu, C. | | Deposit date: | 2021-07-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Two novel human anti-CD25 antibodies with antitumor activity inversely related to their affinity and in vitro activity.
Sci Rep, 11, 2021
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7F0I
 
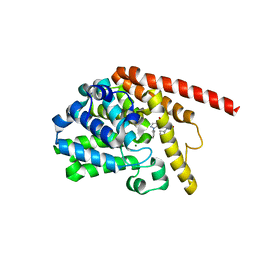 | | phosphodiesterase-9A in complex with inhibitor 4b | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(6-fluoranyl-2-azaspiro[3.3]heptan-2-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, Isoform PDE9A2 of High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.70000887 Å) | | Cite: | Discovery of Potent Phosphodiesterase-9 Inhibitors for the Treatment of Hepatic Fibrosis
J.Med.Chem., 64, 2021
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7QI8
 
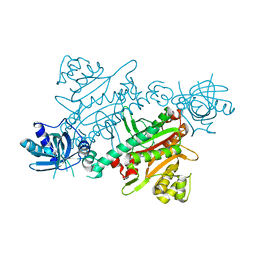 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE AND INHIBITOR | | Descriptor: | 2-azanyl-6-[(1~{S},7~{S})-2,2-bis(fluoranyl)-7-oxidanyl-cycloheptyl]-4-methoxy-7~{H}-pyrrolo[3,4-d]pyrimidin-5-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QHN
 
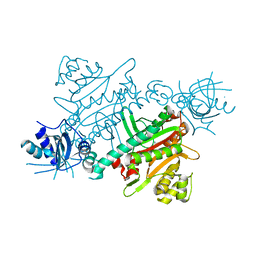 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE and an inhibitor | | Descriptor: | 6-azanyl-2-cyclohexyl-4-fluoranyl-1~{H}-pyrrolo[3,4-c]pyridin-3-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QH8
 
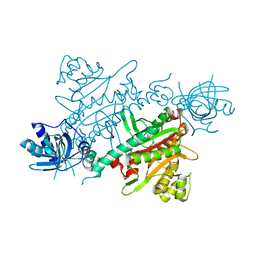 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
2JQC
 
 | | A L-amino acid mutant of a D-amino acid containing conopeptide | | Descriptor: | L-mr12 | | Authors: | Huang, F, Du, W, Han, Y, Wang, C, Chi, C. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Purification and structural characterization of a d-amino acid-containing conopeptide, conomarphin, from Conus marmoreus
Febs J., 275, 2008
|
|
