2J0L
 
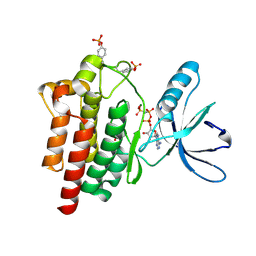 | | Crystal structure of a the active conformation of the kinase domain of focal adhesion kinase with a phosphorylated activation loop. | | Descriptor: | FOCAL ADHESION KINASE 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase
Cell(Cambridge,Mass.), 129, 2007
|
|
8JR9
 
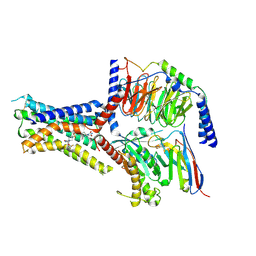 | | Small molecule agonist (PCO371) bound to human parathyroid hormone receptor type 1 (PTH1R) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, He, X, Shan, H, Li, J, Wang, K, Li, Y, Hu, W, Wu, K, Shen, J, Xu, H.E. | | Deposit date: | 2023-06-16 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Conserved class B GPCR activation by a biased intracellular agonist.
Nature, 621, 2023
|
|
2M3E
 
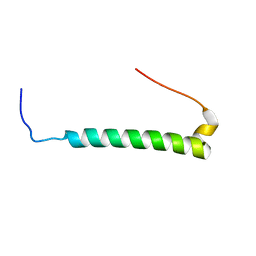 | |
2M6U
 
 | | NMR Structure of CbpAN from Streptococcus pneumoniae | | Descriptor: | Choline binding protein A | | Authors: | Liu, A, Yan, H, Achila, D, Martinez-Hackert, E, Li, Y, Banerjee, R. | | Deposit date: | 2013-04-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Biochem.J., 465, 2015
|
|
2J66
 
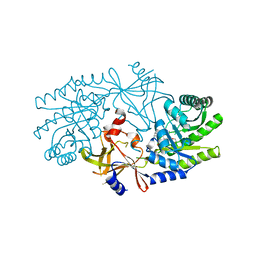 | | Structural characterisation of BtrK decarboxylase from butirosin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, BTRK, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Popovic, B, Li, Y, Chirgadze, D.Y, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2006-09-26 | | Release date: | 2006-09-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterisation of Btrk Decarboxylase from Bacillus Circulans Butirosin Biosynthesis
To be Published
|
|
2ITZ
 
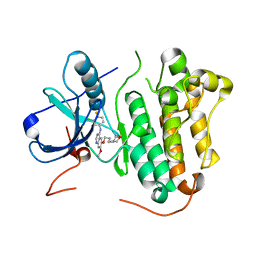 | | Crystal structure of EGFR kinase domain L858R mutation in complex with Iressa | | Descriptor: | CHLORIDE ION, EPIDERMAL GROWTH FACTOR RECEPTOR, Gefitinib | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITX
 
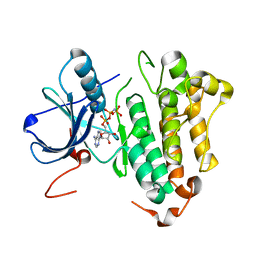 | | Crystal structure of EGFR kinase domain in complex with AMP-PNP | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITY
 
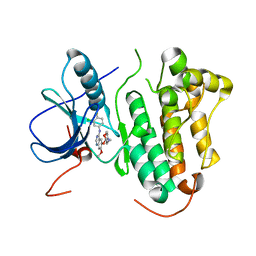 | | Crystal structure of EGFR kinase domain in complex with Iressa | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR, Gefitinib | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITP
 
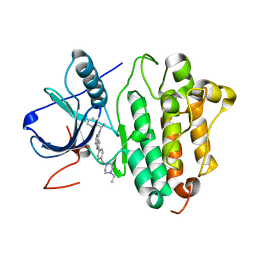 | | Crystal structure of EGFR kinase domain G719S mutation in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR PRECURSOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITW
 
 | | Crystal structure of EGFR kinase domain in complex with AFN941 | | Descriptor: | 1,2,3,4-Tetrahydrogen Staurosporine, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2FMX
 
 | | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+) | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Bian, C, Yuan, C, Li, Y, Huang, M. | | Deposit date: | 2006-01-10 | | Release date: | 2006-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+)
Biochem.Biophys.Res.Commun., 348, 2006
|
|
1WM7
 
 | | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch, 9 structures | | Descriptor: | Neurotoxin BmP01 | | Authors: | Wu, G, Li, Y, Wei, D, He, F, Jiang, S, Hu, G, Wu, H, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 276, 2000
|
|
2G1K
 
 | | Crystal structure of Mycobacterium tuberculosis shikimate kinase in complex with shikimate at 1.75 angstrom resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Gan, J, Gu, Y, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2006-02-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Kinase in Complex with Shikimic Acid and an ATP Analogue.
Biochemistry, 45, 2006
|
|
8EQZ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-C6 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-hexylbenzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2022-10-11 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9F
 
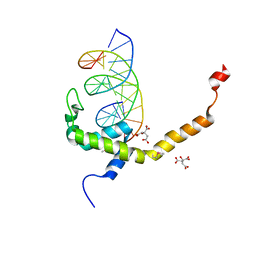 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
6C9J
 
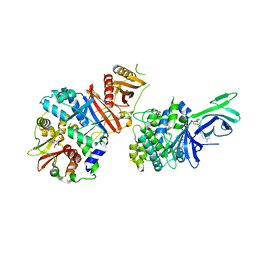 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Brunzelle, J.S, Hitoshi, Y, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6C9F
 
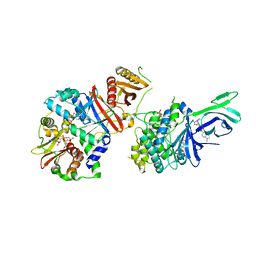 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
2KEO
 
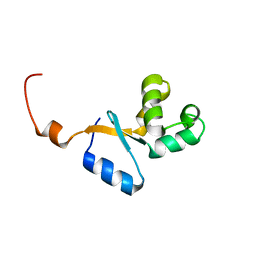 | | Solution NMR structure of human protein HS00059, cytochrome-b5-like domain of the HERC2 E3 ligase. Northeast structural genomics consortium (NESG) target ht98a | | Descriptor: | Probable E3 ubiquitin-protein ligase HERC2 | | Authors: | Lemak, A, Gutmanas, A, Fares, C, Quyang, H, Li, Y, Montelione, G, Arrowsmith, C, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of human protein HS00059
To be Published
|
|
6C9H
 
 | | non-phosphorylated AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
2MQ6
 
 | |
2LGT
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for QFM(Y)F | | Descriptor: | Eukaryotic peptide chain release factor subunit 1 | | Authors: | Wong, L.E, Li, Y, Pillay, S, Pervushin, K. | | Deposit date: | 2011-08-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Selectivity of stop codon recognition in translation termination is modulated by multiple conformations of GTS loop in eRF1
Nucleic Acids Res., 2012
|
|
6C9G
 
 | | AMP-activated protein kinase bound to pharmacological activator R739 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
