4O7X
 
 | |
2H40
 
 | | Crystal structure of the catalytic domain of unliganded PDE5 | | Descriptor: | MAGNESIUM ION, ZINC ION, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Liu, Y, Huai, Q, Cai, J, Zoraghi, R, Francis, S.H, Corbin, J.D, Robinson, H, Xin, Z, Lin, G, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
4NRQ
 
 | | Crystal structure of human ALKBH5 in complex with pyridine-2,4-dicarboxylate | | Descriptor: | MANGANESE (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, RNA demethylase ALKBH5 | | Authors: | Feng, C, Chen, Z, Liu, Y. | | Deposit date: | 2013-11-27 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition
J.Biol.Chem., 289, 2014
|
|
4NRO
 
 | |
4NRM
 
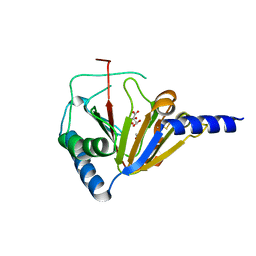 | |
5D84
 
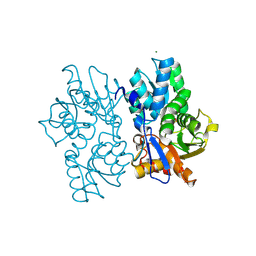 | | Staphyloferrin B precursor biosynthetic enzyme SbnA bound to PLP | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, Probable siderophore biosynthesis protein SbnA | | Authors: | Grigg, J.C, Kobylarz, M.J, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
5E54
 
 | | Two apo structures of the adenine riboswitch aptamer domain determined using an X-ray free electron laser | | Descriptor: | MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
2IS9
 
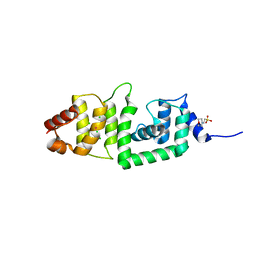 | | Structure of yeast DCN-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Defective in cullin neddylation protein 1, ... | | Authors: | Yang, X, Zhou, J, Sun, L, Wei, Z, Gao, J, Gong, W, Xu, R.M, Rao, Z, Liu, Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for the function of DCN-1 in protein Neddylation.
J.Biol.Chem., 282, 2007
|
|
5EG7
 
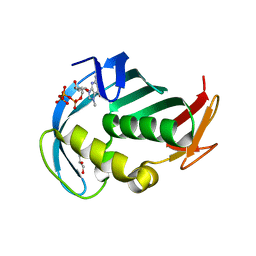 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, Polymerase basic protein 2 | | Authors: | Severin, C, Rocha de Moura, T, Liu, Y, Li, K, Zheng, X, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D87
 
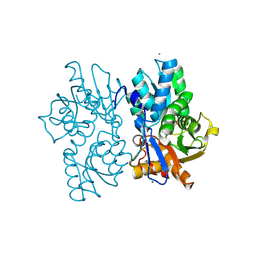 | | Staphyloferrin B precursor biosynthetic enzyme SbnA Y152F/S185G variant | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kobylarz, M.J, Grigg, J.C, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
5D85
 
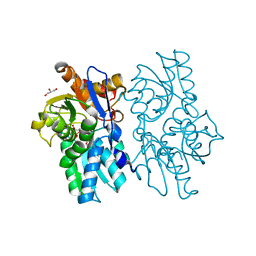 | | Staphyloferrin B precursor biosynthetic enzyme SbnA bound to aminoacrylate intermediate | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CITRATE ANION, GLYCEROL, ... | | Authors: | Kobylarz, M.J, Grigg, J.C, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
5D86
 
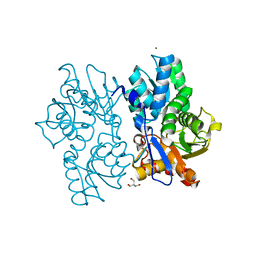 | | Staphyloferrin B precursor biosynthetic enzyme SbnA Y152F variant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kobylarz, M.J, Grigg, J.C, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
2FTP
 
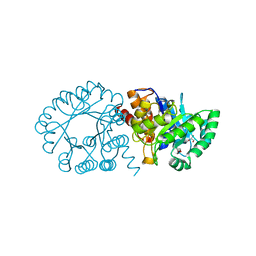 | | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SODIUM ION, hydroxymethylglutaryl-CoA lyase | | Authors: | Xiao, T, Evdokimova, E, Liu, Y, Kudritska, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-24 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa
To be Published
|
|
5EG9
 
 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | Polymerase basic protein 2 | | Authors: | Severin, C, Rocha de Moura, T, Liu, Y, Li, K, Zheng, X, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2J6W
 
 | | R164N mutant of the RUNX1 Runt domain | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Grembecka, J, Zhe, L, Lukasik, S.M, Liu, Y, Bielnicka, I, Bushweller, J.H, Speck, N.A. | | Deposit date: | 2006-10-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mutation in the S-Switch Region of the Runt Domain Alters the Dynamics of an Allosteric Network Responsible for Cbfbeta Regulation.
J.Mol.Biol., 364, 2006
|
|
4QN7
 
 | | Crystal structure of neuramnidase N7 complexed with Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
8AGH
 
 | | BK Polyomavirus VP1 mutant E73A | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
8AGO
 
 | | BK Polyomavirus VP1 mutant E73Q | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
8AH0
 
 | | BK Polyomavirus VP1 mutant VQQ | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
8AH1
 
 | | BK Polyomavirus VP1 mutant N-Q | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
2JVC
 
 | |
4NRP
 
 | |
5EWU
 
 | | Crystal structure of the Arabidopsis thaliana C-terminal Chlh at 1.25A | | Descriptor: | BENZOIC ACID, MAGNESIUM ION, Magnesium-chelatase subunit ChlH, ... | | Authors: | Chen, Z, Zhang, X, Liu, Y, Jiang, L. | | Deposit date: | 2015-11-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the Arabidopsis thaliana C-terminal Chlh at 1.25A
To Be Published
|
|
2HWV
 
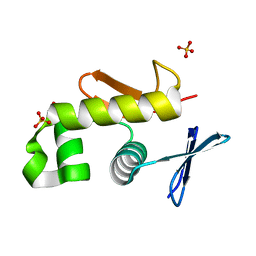 | | Crystal structure of an essential response regulator DNA binding domain, VicRc in Enterococcus faecalis, a member of the YycF subfamily. | | Descriptor: | DNA-binding response regulator VicR, SULFATE ION | | Authors: | Trinh, C.H, Liu, Y, Phillips, S.E.V, Phillips-Jones, M.K. | | Deposit date: | 2006-08-02 | | Release date: | 2007-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the response regulator VicR DNA-binding domain.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4QN5
 
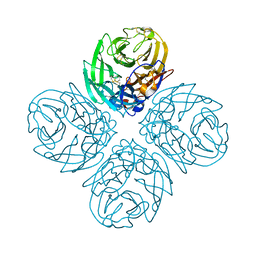 | | Neuraminidase N5 binds LSTa at the second SIA binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
