5ZGC
 
 | | Crystal Structure of SIRT3 in complex with H4K16bhb peptide | | Descriptor: | Histone H4K16bhb peptide, NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2018-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of SIRT3 in complex with H4K16bhb peptide
To be published
|
|
3ZN8
 
 | | Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex | | Descriptor: | 4.5 S RNA, DIPEPTIDYL AMINOPEPTIDASE B, MAGNESIUM ION, ... | | Authors: | von Loeffelholz, O, Knoops, K, Ariosa, A, Zhang, X, Karuppasamy, M, Huard, K, Schoehn, G, Berger, I, Shan, S.O, Schaffitzel, C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural Basis of Signal Sequence Surveillance and Selection by the Srp-Sr Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
6BZE
 
 | | Cryo-EM structure of BCL10 CARD filament | | Descriptor: | B-cell lymphoma/leukemia 10 | | Authors: | David, L, Li, Y, Ma, J, Garner, E, Zhang, X, Wu, H. | | Deposit date: | 2017-12-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Assembly mechanism of the CARMA1-BCL10-MALT1-TRAF6 signalosome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7FBI
 
 | | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5 | | Descriptor: | 3A3 heavy chain, 3A3 light chain, 3A5 heavy chain, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Hong, J, Chen, Y, Zhang, X. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5
To Be Published
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
5WTA
 
 | | Crystal Structure of Staphylococcus aureus SdrE apo form | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
5EH2
 
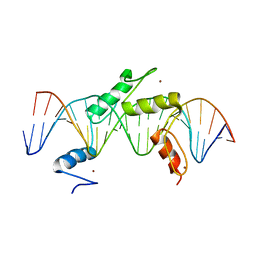 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence III | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EI9
 
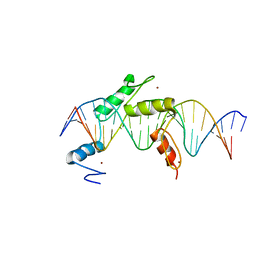 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence I | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*GP*A)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EGB
 
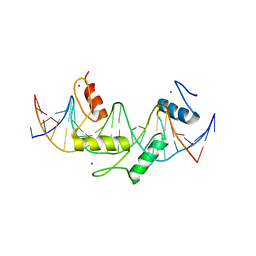 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence II | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
6NT7
 
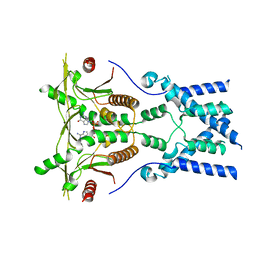 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound dimeric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT8
 
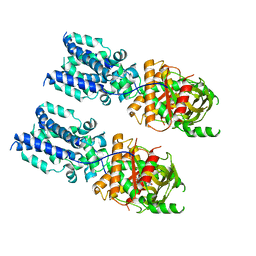 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound tetrameric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT6
 
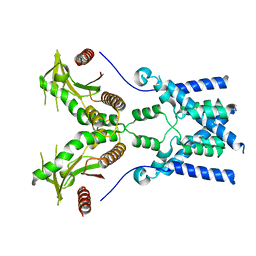 | | Cryo-EM structure of full-length chicken STING in the apo state | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
5WTB
 
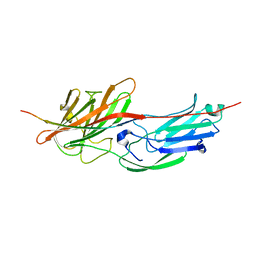 | | Complex Structure of Staphylococcus aureus SdrE with human complement factor H | | Descriptor: | Peptide from Complement factor H, Serine-aspartate repeat-containing protein E | | Authors: | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | Deposit date: | 2016-12-10 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
3UT8
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | IODIDE ION, Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UT7
 
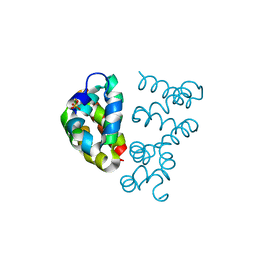 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
8EXE
 
 | |
8EXF
 
 | |
3UT4
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
8I39
 
 | | Cryo-EM structure of abscisic acid transporter AtABCG25 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2023-01-16 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure and molecular mechanism of abscisic acid transporter ABCG25.
Nat.Plants, 9, 2023
|
|
2HMY
 
 | | BINARY COMPLEX OF HHAI METHYLTRANSFERASE WITH ADOMET FORMED IN THE PRESENCE OF A SHORT NONPSECIFIC DNA OLIGONUCLEOTIDE | | Descriptor: | PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), S-ADENOSYLMETHIONINE | | Authors: | O'Gara, M, Zhang, X, Roberts, R.J, Cheng, X. | | Deposit date: | 1999-02-08 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of a binary complex of HhaI methyltransferase with S-adenosyl-L-methionine formed in the presence of a short non-specific DNA oligonucleotide.
J.Mol.Biol., 287, 1999
|
|
2LIR
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in oxidized states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
190D
 
 | |
8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
