3SHS
 
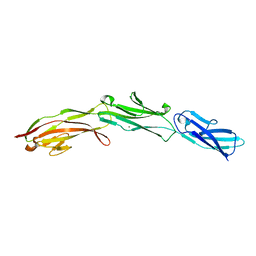 | | Three N-terminal domains of the bacteriophage RB49 Highly Immunogenic Outer Capsid protein (Hoc) | | Descriptor: | Hoc head outer capsid protein, MAGNESIUM ION | | Authors: | Fokine, A, Islam, M.Z, Zhang, Z, Bowman, V.D, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage.
J.Virol., 85, 2011
|
|
5XPZ
 
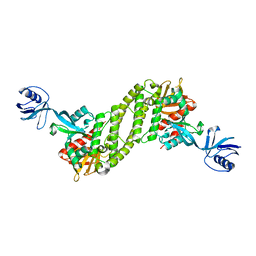 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPY
 
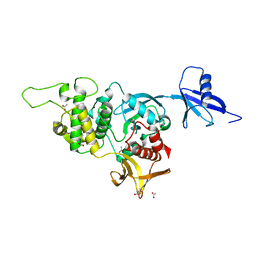 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | ACETATE ION, Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1JOT
 
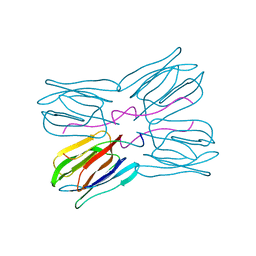 | | STRUCTURE OF THE LECTIN MPA COMPLEXED WITH T-ANTIGEN DISACCHARIDE | | Descriptor: | AGGLUTININ, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Lee, X, Thompson, A, Zhang, Z, Hoa, T.-T, Biesterfeldt, J, Ogata, C, Xu, L, Johnston, R.A.Z, Young, N.M. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of Maclura pomifera agglutinin and the T-antigen disaccharide, Galbeta1,3GalNAc.
J.Biol.Chem., 273, 1998
|
|
5ZMM
 
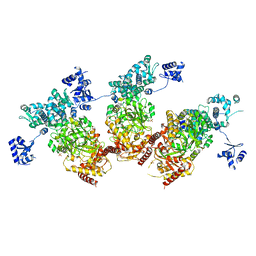 | | Structure of the Type IV phosphorothioation-dependent restriction endonuclease ScoMcrA | | Descriptor: | SULFATE ION, Uncharacterized protein McrA, ZINC ION | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
5ZMN
 
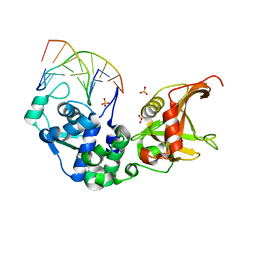 | | Sulfur binding domain and SRA domain of ScoMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*(GS)P*CP*CP*GP*GP*G)-3'), SULFATE ION, Uncharacterized protein McrA | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
2GAS
 
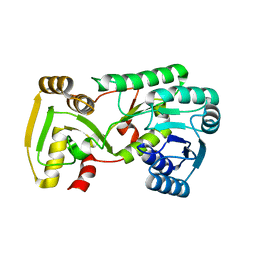 | | Crystal Structure of Isoflavone Reductase | | Descriptor: | isoflavone reductase | | Authors: | Wang, X, He, X, Lin, J, Shao, H, Chang, Z, Dixon, R.A. | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Isoflavone Reductase from Alfalfa (Medicago sativa L.)
J.Mol.Biol., 358, 2006
|
|
4KMA
 
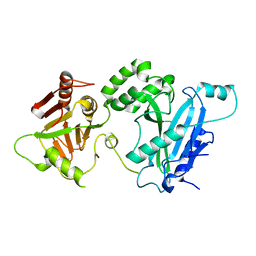 | |
8JIN
 
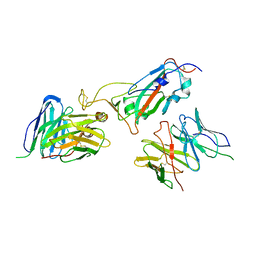 | |
1OPI
 
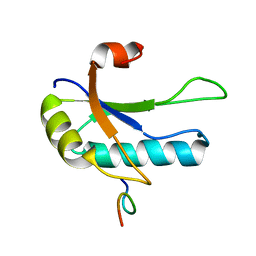 | | SOLUTION STRUCTURE OF THE THIRD RNA RECOGNITION MOTIF (RRM) OF U2AF65 IN COMPLEX WITH AN N-TERMINAL SF1 PEPTIDE | | Descriptor: | SPLICING FACTOR SF1, SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Selenko, P, Gregorovic, G, Sprangers, R, Stier, G, Rhani, Z, Kramer, A, Sattler, M. | | Deposit date: | 2003-03-05 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP
Mol.Cell, 11, 2003
|
|
3ZZM
 
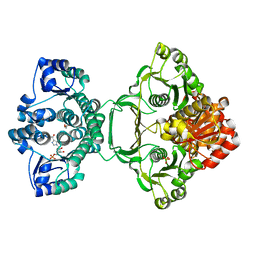 | | Crystal structure of Mycobacterium tuberculosis PurH with a novel bound nucleotide CFAIR, at 2.2 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, GLYCEROL, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-02 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
3J2M
 
 | | The X-ray structure of the gp15 hexamer and the model of the gp18 protein fitted into the cryo-EM reconstruction of the extended T4 tail | | Descriptor: | Tail connector protein Gp15, Tail sheath protein Gp18 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-09 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3J2O
 
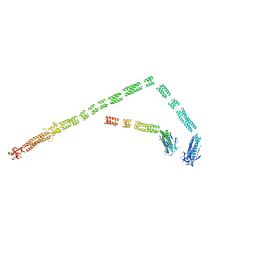 | | Model of the bacteriophage T4 fibritin based on the cryo-EM reconstruction of the contracted T4 tail containing the phage collar and whiskers | | Descriptor: | Fibritin | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A.A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3J2N
 
 | | The X-ray structure of the gp15 hexamer and the model of the gp18 protein fitted into the cryo-EM reconstruction of the contracted T4 tail | | Descriptor: | Tail connector protein Gp15, Tail sheath protein Gp18 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-10 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
4AEZ
 
 | | Crystal Structure of Mitotic Checkpoint Complex | | Descriptor: | MITOTIC SPINDLE CHECKPOINT COMPONENT MAD2, MITOTIC SPINDLE CHECKPOINT COMPONENT MAD3, WD REPEAT-CONTAINING PROTEIN SLP1 | | Authors: | Kulkarni, K.A, Chao, W.C.H, Zhang, Z, Barford, D. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Mitotic Checkpoint Complex
Nature, 484, 2012
|
|
4A2N
 
 | | Crystal Structure of Ma-ICMT | | Descriptor: | CARDIOLIPIN, ISOPRENYLCYSTEINE CARBOXYL METHYLTRANSFERASE, PALMITIC ACID, ... | | Authors: | Yang, J, Kulkarni, K, Manolaridis, I, Zhang, Z, Dodd, R.B, Mas-Droux, C, Barford, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Isoprenylcysteine Carboxyl Methylation from the Crystal Structure of the Integral Membrane Methyltransferase Icmt.
Mol.Cell, 44, 2011
|
|
1O0P
 
 | | Solution Structure of the third RNA Recognition Motif (RRM) of U2AF65 in complex with an N-terminal SF1 peptide | | Descriptor: | Splicing Factor SF1, Splicing factor U2AF 65 kDa subunit | | Authors: | Selenko, P, Gregorovic, G, Sprangers, R, Stier, G, Rhani, Z, Kramer, A, Sattler, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP
Mol.Cell, 11, 2003
|
|
5ZBA
 
 | | Crystal structure of Rtt109-Asf1-H3-H4-CoA complex | | Descriptor: | COENZYME A, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
2ASQ
 
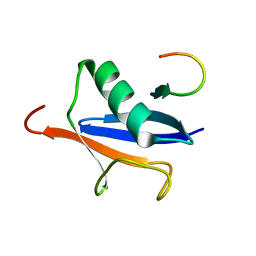 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
4A1O
 
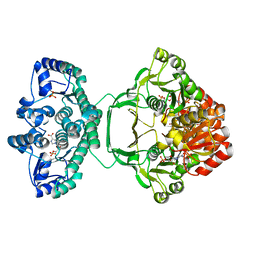 | | Crystal structure of Mycobacterium tuberculosis PurH complexed with AICAR and a novel nucleotide CFAIR, at 2.48 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
1XI2
 
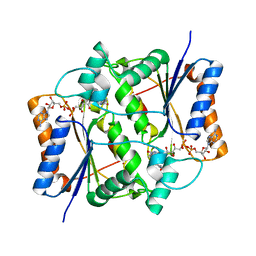 | | Quinone Reductase 2 in Complex with Cancer Prodrug CB1954 | | Descriptor: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, NRH dehydrogenase [quinone] 2, ... | | Authors: | Fu, Y, Buryanovskyy, L, Zhang, Z. | | Deposit date: | 2004-09-21 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of quinone reductase 2 in complex with cancer prodrug CB1954
Biochem.Biophys.Res.Commun., 336, 2005
|
|
5ZB9
 
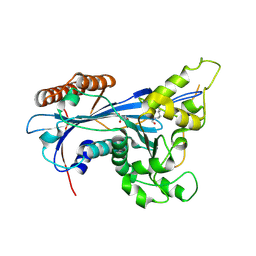 | | Crystal structure of Rtt109 from Aspergillus fumigatus | | Descriptor: | DNA damage response protein Rtt109, putative, GLYCEROL | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
5XRF
 
 | | Crystal structure of Da-36, a thrombin-like enzyme from Deinagkistrodon acutus | | Descriptor: | DI(HYDROXYETHYL)ETHER, Snake venom serine protease Da-36, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, C.-C, Wu, W.-G, Fan, Q, Tian, J, Zhang, Z, Zheng, Y. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Da-36, a thrombin-like enzyme from Deinagkistrodon acutus
To Be Published
|
|
2VOK
 
 | | Murine TRIM21 | | Descriptor: | 52 KDA RO PROTEIN | | Authors: | Keeble, A.H, Khan, Z, Forster, A, James, L.C. | | Deposit date: | 2008-02-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Trim21 is an Igg Receptor that is Structurally, Thermodynamically, and Kinetically Conserved.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
