1YD7
 
 | | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha, UNKNOWN ATOM OR ION | | Authors: | Horanyi, P, Florence, Q, Zhou, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus
To be published
|
|
1YCY
 
 | | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein | | Authors: | Huang, L, Liu, Z.-J, Lee, D, Tempel, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conserved hypothetical protein Pfu-1806301-001 from Pyrococcus furiosus
To be published
|
|
1Y81
 
 | | Conserved hypothetical protein Pfu-723267-001 from Pyrococcus furiosus | | Descriptor: | COENZYME A, THIOCYANATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Zhao, M, Chang, J, Habel, J, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Tempel, W, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Conserved hypothetical protein Pfu-723267-001 from Pyrococcus furiosus
To be published
|
|
5VIO
 
 | | Crystal structure of ASK1 kinase domain with a potent inhibitor (analog 13) | | Descriptor: | 4-methoxy-N~1~-methyl-N~3~-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}benzene-1,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Jasti, J, Chang, J, Kurumbail, R. | | Deposit date: | 2017-04-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Rational approach to highly potent and selective apoptosis signal-regulating kinase 1 (ASK1) inhibitors.
Eur J Med Chem, 145, 2017
|
|
8TOB
 
 | | Acinetobacter GP16 Type IV pilus | | Descriptor: | Fimbrial protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TWC
 
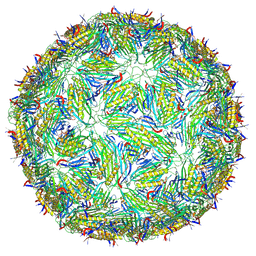 | | Acinetobacter phage AP205 T=3 VLP | | Descriptor: | Coat protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TV9
 
 | | Inner Mat-T4P complex | | Descriptor: | Fimbrial protein, Maturation protein | | Authors: | Meng, R, Xing, Z, Thongchol, J, Zhang, J. | | Deposit date: | 2023-08-17 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (8.15 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TVA
 
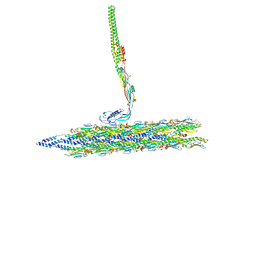 | | Outer Mat-T4P complex | | Descriptor: | Fimbrial protein, Maturation protein | | Authors: | Meng, R, Xing, Z, Thongchol, J, Zhang, J. | | Deposit date: | 2023-08-17 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
8TW2
 
 | | Acinetobacter phage AP205 T=4 VLP | | Descriptor: | Coat protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
1YBY
 
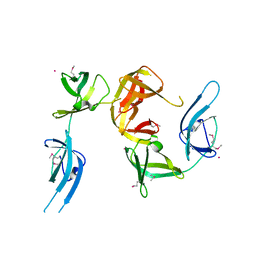 | | Conserved hypothetical protein Cth-95 from Clostridium thermocellum | | Descriptor: | Translation elongation factor P, UNKNOWN ATOM OR ION | | Authors: | Zhao, M, Zhou, W, Chang, J, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Tempel, W, Lin, D, Zhang, H, Ljundahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conserved hypothetical protein Cth-95 from Clostridium thermocellum
To be published
|
|
7O1N
 
 | |
7OMK
 
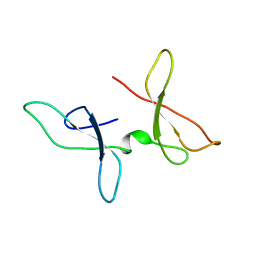 | | The NMR structure of the Zf-GRF domains from the mouse Endonuclease VIII-LIKE 3 (mNEIL3) | | Descriptor: | Endonuclease 8-like 3 | | Authors: | Dinesh, D.C, Huskova, A, Srb, P, Veverka, V, Silhan, J. | | Deposit date: | 2021-05-24 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Model of abasic site DNA cross-link repair; from the architecture of NEIL3 DNA binding domains to the X-structure model.
Nucleic Acids Res., 50, 2022
|
|
5IZ8
 
 | | Protein-protein interaction | | Descriptor: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ9
 
 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZA
 
 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
6PMO
 
 | |
5IZ6
 
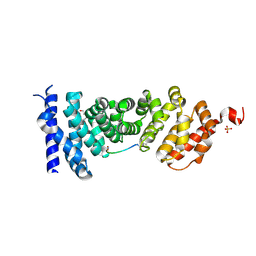 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5B6G
 
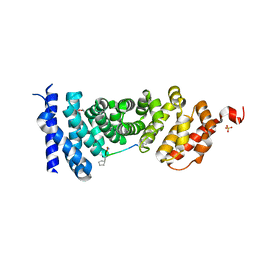 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
6WXK
 
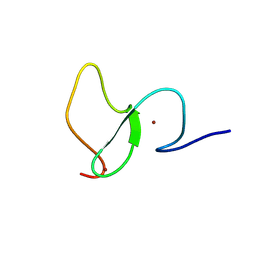 | | PHF23 PHD Domain Apo | | Descriptor: | PHD finger protein 23, ZINC ION | | Authors: | Vann, K.R, Zhang, J, Zhang, Y, Kutateladze, T. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion.
Nat Commun, 11, 2020
|
|
5XJC
 
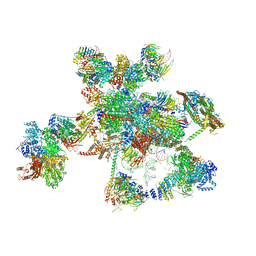 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | Deposit date: | 2017-04-30 | | Release date: | 2017-07-05 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
5MLB
 
 | | Crystal structure of human RAS in complex with darpin K27 | | Descriptor: | DARPin K27, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Overman, R, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | INhibition of RAS nucleotide exchange by a DARPin: structural characterisation and effects on downstream signalling by active RAS
To Be Published
|
|
8H91
 
 | |
8HQG
 
 | |
5MLA
 
 | | Crystal structure of human RAS in complex with darpin K55 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Overman, R, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Inhibition of RAS nucleotide exchange by a DARPin: structural characterisation and effects on downstream signalling by active RAS
To Be Published
|
|
8HVM
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF07321332
To Be Published
|
|
