5GRM
 
 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
7LUH
 
 | |
7LUJ
 
 | |
7LO1
 
 | |
7LDF
 
 | |
7M2G
 
 | | INTERLEUKIN-2 (human) mutant P65K, C125S | | Descriptor: | GLYCEROL, Interleukin-2, SULFATE ION | | Authors: | Ptacin, J.L, Milla, M.E, Steinbacher, S, Maskos, K, Thomsen, M. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An engineered IL-2 reprogrammed for anti-tumor therapy using a semi-synthetic organism.
Nat Commun, 12, 2021
|
|
7MD2
 
 | | The F1 region of ammocidin-bound Saccharomyces cerevisiae ATP synthase | | Descriptor: | (3~{E},5~{Z},7~{E},9~{R},10~{S},11~{E},13~{E},15~{E},17~{R},18~{S},20~{S})-20-[(1~{R})-1-[(2~{S},3~{R},4~{R},5~{S},6~{R})-5-[(2~{S},4~{S},5~{S},6~{R})-5-[(2~{S},4~{R},5~{R},6~{R})-4,6-dimethyl-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-4-oxidanyl-oxan-2-yl]oxy-3-methoxy-6-(3-methoxypropyl)-5-methyl-2,4-bis(oxidanyl)oxan-2-yl]ethyl]-5,18-dimethoxy-3,7,9,11,13,15-hexamethyl-10-[(2~{R},3~{S},4~{R},5~{R},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-17-oxidanyl-1-oxacycloicosa-3,5,7,11,13,15-hexaen-2-one, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2021-04-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A Family of Glycosylated Macrolides Selectively Target Energetic Vulnerabilities in Leukemia
Biorxiv, 2021
|
|
7MD3
 
 | | The F1 region of apoptolidin-bound Saccharomyces cerevisiae ATP synthase | | Descriptor: | (3~{E},5~{E},7~{E},9~{R},10~{R},11~{E},13~{E},17~{S},18~{S},20~{S})-18-methoxy-20-[(~{R})-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-[(2~{R})-3-methoxy-2-[(2~{R},4~{S},5~{S},6~{S})-5-[(2~{S},4~{R},5~{R},6~{R})-4-methoxy-6-methyl-5-oxidanyl-oxan-2-yl]oxy-4,6-dimethyl-4-oxidanyl-oxan-2-yl]oxy-propyl]-3,5-dimethyl-2,4-bis(oxidanyl)oxan-2-yl]-oxidanyl-methyl]-10-[(2~{R},3~{S},4~{S},5~{R},6~{S})-5-methoxy-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-3,5,7,9,13-pentamethyl-17-oxidanyl-1-oxacycloicosa-3,5,7,11,13-pentaen-2-one, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2021-04-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A Family of Glycosylated Macrolides Selectively Target Energetic Vulnerabilities in Leukemia
Biorxiv, 2021
|
|
5GS5
 
 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
7M7C
 
 | | Crystal Structure of Hip1 (Rv2224c) mutant - T466A/S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors and Inactivators of Mycobacterium tuberculosis serine protease Hip1 (Rv2224c)
To Be Published
|
|
1GCD
 
 | |
1GMH
 
 | |
1GQR
 
 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH RIVASTIGMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(1S)-1-(DIMETHYLAMINO)ETHYL]PHENOL, ACETYLCHOLINESTERASE, ... | | Authors: | Raves, M.L, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Structural Studies on the Interaction of Cholinesterases with the Anti-Alzheimer Drug Rivastigmine
Biochemistry, 41, 2002
|
|
1HCZ
 
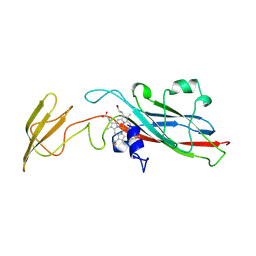 | | LUMEN-SIDE DOMAIN OF REDUCED CYTOCHROME F AT-35 DEGREES CELSIUS | | Descriptor: | CYTOCHROME F, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain.
Protein Sci., 5, 1996
|
|
1GQS
 
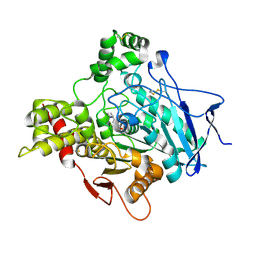 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH NAP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(1S)-1-(DIMETHYLAMINO)ETHYL]PHENOL, ACETYLCHOLINESTERASE | | Authors: | Bar-on, P, Millard, C.B, Harel, M, Dvir, H, Enz, A, Sussman, J.L, Silman, I. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetic and Structural Studies on the Interaction of Cholinesterases with the Anti-Alzheimer Drug Rivastigmine
Biochemistry, 41, 2002
|
|
7MPE
 
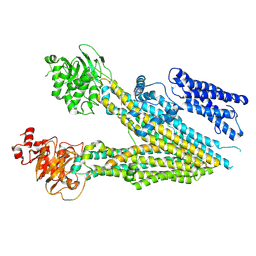 | |
5ICD
 
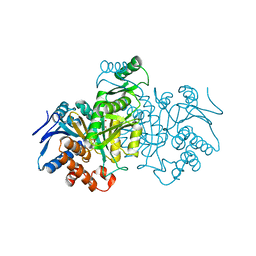 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
7LG6
 
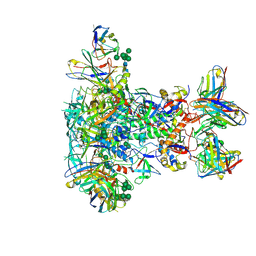 | | BG505 SOSIP.v5.2 in complex with VRC40.01 and RM19R Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Torres, J.L, Wu, N.R, Ward, A.B. | | Deposit date: | 2021-01-19 | | Release date: | 2021-09-15 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies.
Cell Rep, 37, 2021
|
|
1L7F
 
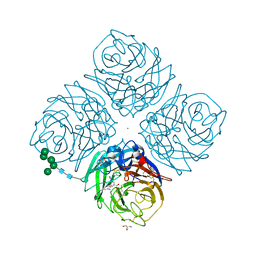 | | Crystal structure of influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
7L99
 
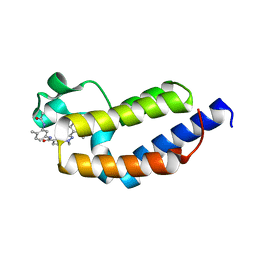 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1302 | | Descriptor: | Bromodomain testis-specific protein, N-[3-(acetylamino)-4-methylphenyl]-3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carboxamide, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Sharma, R, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LKF
 
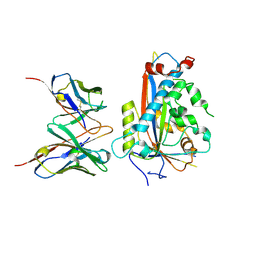 | | WT Chicken Scap L1-L7 / Fab 4G10 complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4G10 heavy chain, 4G10 light chain, ... | | Authors: | Kober, D.L, Radhakrishnan, A, Goldstein, J.L, Brown, M.S, Clark, L.D, Bai, X.-C, Rosenbaum, D.M. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing.
Cell, 184, 2021
|
|
7L9A
 
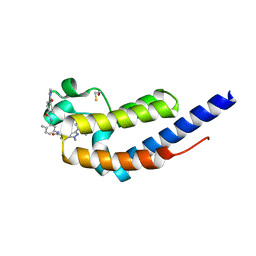 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1102 | | Descriptor: | BETA-MERCAPTOETHANOL, Bromodomain testis-specific protein, N~1~-(5-{[3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carbonyl]amino}-2-methylphenyl)-N~4~-methylbenzene-1,4-dicarboxamide | | Authors: | Sharma, R, Kaur, G, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LKH
 
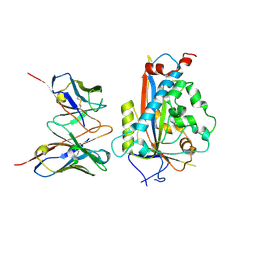 | | Chicken Scap D435V L1-L7 domain / Fab complex focused map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4G10 Fab heavy chain, 4G10 Fab kappa chain, ... | | Authors: | Kober, D.L, Radhakrishnan, A, Goldstein, J.L, Brown, M.S, Clark, L.D, Bai, X.-C, Rosenbaum, D.M. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing.
Cell, 184, 2021
|
|
7NVH
 
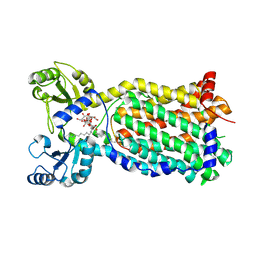 | | Cryo-EM structure of the mycolic acid transporter MmpL3 from M. tuberculosis | | Descriptor: | 2-decyl-2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}dodecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Trehalose monomycolate exporter MmpL3 | | Authors: | Adams, O, Deme, J.C, Parker, J.L, Lea, S.M, Newstead, S. | | Deposit date: | 2021-03-15 | | Release date: | 2021-06-16 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure and resistance landscape of M. tuberculosis MmpL3: An emergent therapeutic target.
Structure, 29, 2021
|
|
7NQK
 
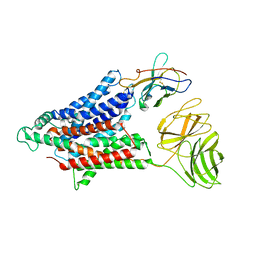 | | Cryo-EM structure of the mammalian peptide transporter PepT2 | | Descriptor: | Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-03-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of PepT2 reveals structural basis for proton-coupled peptide and prodrug transport in mammals.
Sci Adv, 7, 2021
|
|
