5J6F
 
 | | Crystal structure of DAH7PS-CM complex from Geobacillus sp. with prephenate | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, chorismate mutase-isozyme 3, MANGANESE (II) ION, ... | | Authors: | Nazmi, A.R, Othman, M, Lang, E.J.M, Bai, Y, Allison, T.M, Panjkar, S, Arcus, V.L, Parker, E.J. | | Deposit date: | 2016-04-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Interdomain Conformational Changes Provide Allosteric Regulation en Route to Chorismate.
J. Biol. Chem., 291, 2016
|
|
5CIL
 
 | | Crystal Structure of non-neutralizing version of 4E10 (WDWD) with epitope bound | | Descriptor: | ACETATE ION, CHLORIDE ION, FAB 4E10 HEAVY CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
5CIP
 
 | | Crystal Structure of Unbound 4E10 | | Descriptor: | FAB 4E10 HEAVY CHAIN, FAB 4E10 LIGHT CHAIN | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
5CIN
 
 | | Crystal Structure of non-neutralizing version of 4E10 (DeltaLoop) with epitope bound | | Descriptor: | CHLORIDE ION, FAB 4E10 HEAVY CHAIN, FAB 4E10 LIGHT CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
5JYM
 
 | | Human P-cadherin EC12 with scFv TSP11 bound | | Descriptor: | Antibody scFv TSP11, CALCIUM ION, Cadherin-3 | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Disruption of cell adhesion by an antibody targeting the cell-adhesive intermediate (X-dimer) of human P-cadherin
Sci Rep, 7, 2017
|
|
5JYL
 
 | | Human P-cadherin MEC1 with scFv TSP7 bound | | Descriptor: | CHLORIDE ION, Cadherin-3, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Disruption of cell adhesion by an antibody targeting the cell-adhesive intermediate (X-dimer) of human P-cadherin
Sci Rep, 7, 2017
|
|
4KS2
 
 | | Influenza Neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-carbamimidamido-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-carbamimidamido-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
3QUG
 
 | | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Gallium-porphyrin | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING GA, ... | | Authors: | Moriwaki, Y, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2011-02-24 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of recognition of antibacterial porphyrins by heme-transporter IsdH-NEAT3 of Staphylococcus aureus.
Biochemistry, 50, 2011
|
|
7OMS
 
 | | Bs164 in complex with mannocyclophellitol aziridine | | Descriptor: | (1~{R},2~{R},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | McGregor, N, Beenakker, T, Kuo, C.L, Wong, C.S, Offren, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H, Davies, G.J. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
3QUH
 
 | |
4A1O
 
 | | Crystal structure of Mycobacterium tuberculosis PurH complexed with AICAR and a novel nucleotide CFAIR, at 2.48 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
3ZZM
 
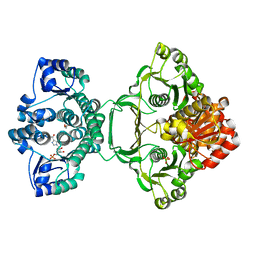 | | Crystal structure of Mycobacterium tuberculosis PurH with a novel bound nucleotide CFAIR, at 2.2 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, GLYCEROL, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-02 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
7SGD
 
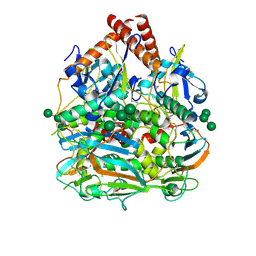 | | Lassa virus glycoprotein construct(Josiah GPCysR4) recovered from GPC-I53-50 nanoparticle by localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Antanasijevic, A, Brouwer, P.J.M, Ward, A.B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-12 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection.
Cell Host Microbe, 30, 2022
|
|
2VW1
 
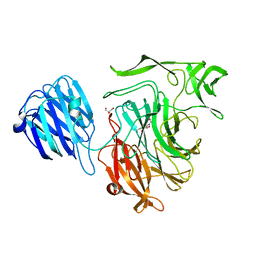 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2X2T
 
 | | CRYSTAL STRUCTURE OF SCLEROTINIA SCLEROTIORUM AGGLUTININ SSA in complex with Gal-beta1,3-Galnac | | Descriptor: | AGGLUTININ, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Rouge, P, Van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2010-01-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of the Galnac/Gal-Specific Agglutinin from the Phytopathogenic Ascomycete Sclerotinia Sclerotiorum Reveals Novel Adaptation of a Beta-Trefoil Domain
J.Mol.Biol., 400, 2010
|
|
2X2S
 
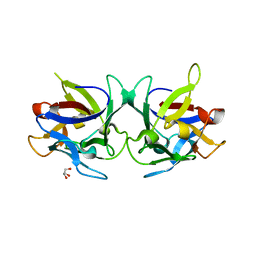 | | Crystal structure of Sclerotinia sclerotiorum agglutinin SSA | | Descriptor: | AGGLUTININ, GLYCEROL | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Rouge, P, Van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2010-01-15 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Galnac/Gal-Specific Agglutinin from the Phytopathogenic Ascomycete Sclerotinia Sclerotiorum Reveals Novel Adaptation of a Beta-Trefoil Domain
J.Mol.Biol., 400, 2010
|
|
7TIO
 
 | |
7TIR
 
 | |
7TIP
 
 | |
7TIS
 
 | |
7TIQ
 
 | |
2VW2
 
 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2VW0
 
 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2GRW
 
 | | Solution structure of the poliovirus 3'-UTR Y-stem | | Descriptor: | 5'-GGACCUCUCGAAAGAGAUGUCC-3' | | Authors: | Heus, H.A, Zoll, J, Tessari, M, van Kuppeveld, F.J.M, Melchers, W.J.G. | | Deposit date: | 2006-04-25 | | Release date: | 2007-03-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Breaking pseudo-twofold symmetry in the poliovirus 3'-UTR Y-stem by restoring Watson-Crick base pairs.
Rna, 13, 2007
|
|
7URJ
 
 | |
