3AGG
 
 | | X-ray analysis of lysozyme in the absence of Arg | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
3AGH
 
 | | X-ray analysis of lysozyme in the presence of 200 mM Arg | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
3AGI
 
 | | High resolution X-ray analysis of Arg-lysozyme complex in the presence of 500 mM Arg | | Descriptor: | ACETATE ION, ARGININE, CHLORIDE ION, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
7VDN
 
 | | High resolution crystal structure of Sperm Whale Myoglobin in the carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shibayama, N, Sato-Tomita, A, Ishimoto, N, Park, S.Y. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | X-ray fluorescence holography of biological metal sites: Application to myoglobin.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
7Y4A
 
 | | Crystal structure of human ELMO1 RBD-RhoG complex | | Descriptor: | Engulfment and cell motility protein 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tsuda, K, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Targeting Ras-binding domain of ELMO1 by computational nanobody design.
Commun Biol, 6, 2023
|
|
1LUE
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68A/D122N MUTANT (MET) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-05-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
3A4P
 
 | | human c-MET kinase domain complexed with 6-benzyloxyquinoline inhibitor | | Descriptor: | (2E)-3-{6-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]quinolin-3-yl}-N-methylprop-2-enamide, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Fukami, T.A, Kadono, S, Yamamuro, M, Matsuura, T. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery of 6-benzyloxyquinolines as c-Met selective kinase inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5XZV
 
 | | Crystal structure of Rad53 1-466 in complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase RAD53 | | Authors: | Weng, J.H, Tsai, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phospho-Priming Confers Functionally Relevant Specificities for Rad53 Kinase Autophosphorylation
Biochemistry, 56, 2017
|
|
5XZW
 
 | | Crystal structure of Rad53 1-466 | | Descriptor: | Serine/threonine-protein kinase RAD53 | | Authors: | Weng, J.H, Tsai, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phospho-Priming Confers Functionally Relevant Specificities for Rad53 Kinase Autophosphorylation
Biochemistry, 56, 2017
|
|
3WZD
 
 | | KDR in complex with ligand lenvatinib | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-{3-chloro-4-[(cyclopropylcarbamoyl)amino]phenoxy}-7-methoxyquinoline-6-carboxamide, ... | | Authors: | Okamoto, K, Ikemori_Kawada, M, Inoue, A, Matsui, J. | | Deposit date: | 2014-09-24 | | Release date: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Distinct binding mode of multikinase inhibitor lenvatinib revealed by biochemical characterization.
ACS MED.CHEM.LETT., 6, 2015
|
|
3WZE
 
 | | KDR in complex with ligand sorafenib | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, ACETATE ION, ... | | Authors: | Okamoto, K, Ikemori_Kawada, M, Inoue, A, Matsui, J. | | Deposit date: | 2014-09-24 | | Release date: | 2015-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct binding mode of multikinase inhibitor lenvatinib revealed by biochemical characterization.
ACS MED.CHEM.LETT., 6, 2015
|
|
2QYF
 
 | |
2CU7
 
 | | Solution structure of the SANT domain of human KIAA1915 protein | | Descriptor: | KIAA1915 protein | | Authors: | Yoneyama, M, Umehara, T, Saito, K, Tochio, N, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-25 | | Release date: | 2005-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
2CUJ
 
 | | Solution structure of SWIRM domain of mouse transcriptional adaptor 2-like | | Descriptor: | transcriptional adaptor 2-like | | Authors: | Yoneyama, M, Umehara, T, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
2DCE
 
 | | Solution structure of the SWIRM domain of human KIAA1915 protein | | Descriptor: | KIAA1915 protein | | Authors: | Yoneyama, M, Tochio, N, Umehara, T, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-06 | | Release date: | 2006-07-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
2D3Q
 
 | |
3AGW
 
 | |
2E5S
 
 | | Solution structure of the zf-CCCHx2 domain of muscleblind-like 2, isoform 1 [Homo sapiens] | | Descriptor: | OTTHUMP00000018578, ZINC ION | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
2E61
 
 | | Solution structure of the zf-CW domain in zinc finger CW-type PWWP domain protein 1 | | Descriptor: | ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
3AUW
 
 | |
2D11
 
 | |
2DBR
 
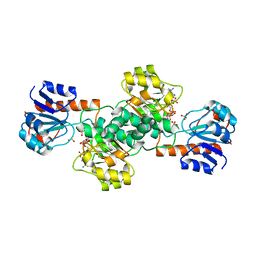 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (P1) | | Descriptor: | Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DBQ
 
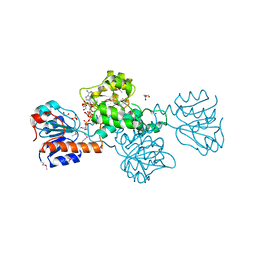 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (I41) | | Descriptor: | GLYCEROL, Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DBZ
 
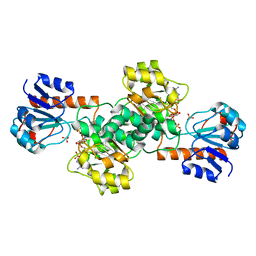 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (P61) | | Descriptor: | Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DGA
 
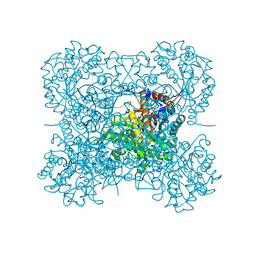 | | Crystal structure of hexameric beta-glucosidase in wheat | | Descriptor: | Beta-glucosidase, GLYCEROL, SULFATE ION | | Authors: | Sue, M, Yamazaki, K, Miyamoto, T, Yajima, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and Structural Characterization of Hexameric beta-D-Glucosidases in Wheat and Rye.
Plant Physiol., 141, 2006
|
|
