8I7Z
 
 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B5 | | Descriptor: | 5-[3-(2-methoxy-5-oxidanyl-phenyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N,N-dimethyl-pyridine-3-carboxamide, BETA-ALANINE, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25495815 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
8I7T
 
 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B4 | | Descriptor: | Tyrosine-protein kinase ABL1, [3-[5-[5-(dimethylcarbamoyl)pyridin-3-yl]-1H-pyrrolo[2,3-b]pyridin-3-yl]-4-methoxy-phenyl] ethanesulfonate | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80004954 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
7VBT
 
 | | Crystal structure of RIOK2 in complex with CQ211 | | Descriptor: | 8-(6-methoxypyridin-3-yl)-1-[4-piperazin-1-yl-3-(trifluoromethyl)phenyl]-5H-[1,2,3]triazolo[4,5-c]quinolin-4-one, Serine/threonine-protein kinase RIO2 | | Authors: | Zhu, C, Zhang, Z.M. | | Deposit date: | 2021-09-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54001474 Å) | | Cite: | Discovery of 8-(6-Methoxypyridin-3-yl)-1-(4-(piperazin-1-yl)-3-(trifluoromethyl)phenyl)-1,5-dihydro- 4H -[1,2,3]triazolo[4,5- c ]quinolin-4-one (CQ211) as a Highly Potent and Selective RIOK2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
7W7Y
 
 | | The crystal structure of human abl1 kinase domain in complex with ABL2-A5 | | Descriptor: | 5-[3-(5-methanoyl-2-methoxy-4-oxidanyl-phenyl)-1~{H}-pyrrolo[2,3-b]pyridin-5-yl]-~{N},~{N}-dimethyl-pyridine-3-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z.M. | | Deposit date: | 2021-12-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.20003033 Å) | | Cite: | Cell-Active, Reversible, and Irreversible Covalent Inhibitors That Selectively Target the Catalytic Lysine of BCR-ABL Kinase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7W7X
 
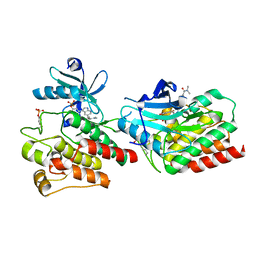 | | The crystal structure of human abl1 kinase domain in complex with ABL1-A11 | | Descriptor: | 5-[5-(dimethylcarbamoyl)pyridin-3-yl]-3-(5-fluorosulfonyloxy-2-methoxy-phenyl)-1H-pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z.M. | | Deposit date: | 2021-12-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.0000093 Å) | | Cite: | Cell-Active, Reversible, and Irreversible Covalent Inhibitors That Selectively Target the Catalytic Lysine of BCR-ABL Kinase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1XBT
 
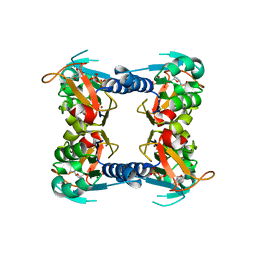 | | Crystal Structure of Human Thymidine Kinase 1 | | Descriptor: | MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE, Thymidine kinase, ... | | Authors: | Welin, M, Kosinska, U, Mikkelsen, N.E, Carnrot, C, Zhu, C, Wang, L, Eriksson, S, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2004-08-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of thymidine kinase 1 of human and mycoplasmic origin
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4ZDS
 
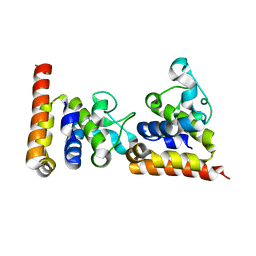 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | Descriptor: | Protein ETHYLENE INSENSITIVE 3 | | Authors: | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | Deposit date: | 2015-04-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
5K02
 
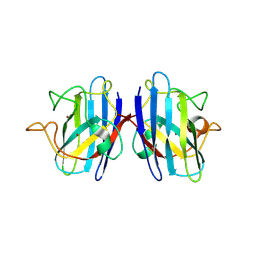 | | Structure of human SOD1 with T2D mutation | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Fay, J.M, Zhu, C, Cui, W, Ke, H, Dokholyan, N.V. | | Deposit date: | 2016-05-17 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Phosphomimetic Mutation Stabilizes SOD1 and Rescues Cell Viability in the Context of an ALS-Associated Mutation.
Structure, 24, 2016
|
|
4F5W
 
 | | Crystal structure of ligand free human STING CTD | | Descriptor: | CALCIUM ION, Transmembrane protein 173 | | Authors: | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
4F5Y
 
 | | Crystal structure of human STING CTD complex with C-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TEF
 
 | | Crystal Structure of the Periplasmic Catecholate-Siderophore Binding Protein VctP from Vibrio Cholerae | | Descriptor: | Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | Authors: | Liu, X, Wang, Z, Liu, S, Li, N, Chen, Y, Zhu, C, Zhu, D, Wei, T, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-13 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of periplasmic catecholate-siderophore binding protein VctP from Vibrio cholerae at 1.7 A resolution
Febs Lett., 586, 2012
|
|
2UZ3
 
 | | Crystal Structure of Thymidine Kinase with dTTP from U. urealyticum | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Welin, M, Kosinska, U, Mikkelsen, N.E, Carnrot, C, Zhu, C, Wang, L, Eriksson, S, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2007-04-24 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Thymidine Kinase 1 of Human and Mycoplasmic Origin
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8HGI
 
 | | Crystal structure of VNAR aGFP14 in complex with GFP | | Descriptor: | GFP, VNAR aGFP14 | | Authors: | Zheng, P, Zhu, C, Jin, T. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selection, identification and crystal structure of shark-derived single-domain antibodies against a green fluorescent protein.
Int.J.Biol.Macromol., 247, 2023
|
|
5ZC3
 
 | | The Crystal Structure of PcRxLR12 | | Descriptor: | RxLR effector | | Authors: | Zhao, L, Zhang, X, Zhu, C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Crystal structure of the RxLR effector PcRxLR12 from Phytophthora capsici
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5X74
 
 | | The crystal Structure PDE delta in complex with compound (R, R)-1g | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-[2-[4-[(2R)-2-(2-fluorophenyl)-4-oxidanylidene-1,2-dihydroquinazolin-3-yl]piperidin-1-yl]ethyl]-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5X73
 
 | | The crystal Structure PDE delta in complex with R-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
5X72
 
 | | The crystal Structure PDE delta in complex with (rac)-p9 | | Descriptor: | (2R)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, (2S)-2-(2-fluorophenyl)-3-phenyl-1,2-dihydroquinazolin-4-one, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Jiang, Y, Zhuang, C, Chen, L, Wang, R, Wang, F, Sheng, C. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Biology-Inspired Discovery of Novel KRAS-PDE delta Inhibitors
J. Med. Chem., 60, 2017
|
|
7F09
 
 | | Crystal structure of the HLH-Lz domain of human TFE3 | | Descriptor: | 1,2-ETHANEDIOL, Transcription factor E3, ZINC ION | | Authors: | Yang, G, Li, P, Liu, Z, Wu, S, Zhuang, C, Qiao, H, Fang, P, Wang, J. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the dimerization mechanism of human transcription factor E3.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
4NII
 
 | |
4NIH
 
 | |
4NIG
 
 | |
4NID
 
 | | Crystal structure of AlkB protein with cofactors bound to dsDNA containing m6A | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*AP*AP*CP*GP*GP*TP*AP*TP*TP*AP*CP*CP*T)-3', 5'-D(*TP*AP*GP*GP*TP*AP*AP*(6MA)P*AP*CP*CP*GP*T)-3', ... | | Authors: | Zhu, C.X, Yi, C.Q. | | Deposit date: | 2013-11-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Switching Demethylation Activities between AlkB Family RNA/DNA Demethylases through Exchange of Active-Site Residues.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7C4D
 
 | | Marine microorganism esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative esterase, ... | | Authors: | Zhu, C.H, Wu, Y.K, Isupov, M.N. | | Deposit date: | 2020-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into a Novel Esterase from the East Pacific Rise and Its Improved Thermostability by a Semirational Design.
J.Agric.Food Chem., 69, 2021
|
|
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | Authors: | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-04-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7BZF
 
 | | COVID-19 RNA-dependent RNA polymerase post-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (31-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-04-27 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
