2NR2
 
 | | The MUMO (minimal under-restraining minimal over-restraining) method for the determination of native states ensembles of proteins | | Descriptor: | Ubiquitin | | Authors: | Richter, B, Gsponer, J, Varnai, P, Salvatella, X, Vendruscolo, M. | | Deposit date: | 2006-11-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The MUMO (minimal under-restraining minimal over-restraining) method for the determination of native state ensembles of proteins
J.Biomol.Nmr, 37, 2007
|
|
2LP5
 
 | | Native Structure of the Fyn SH3 A39V/N53P/V55L | | Descriptor: | Tyrosine-protein kinase Fyn | | Authors: | Neudecker, P, Robustelli, P, Cavalli, A, Vendruscolo, M, Kay, L.E. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an intermediate state in protein folding and aggregation.
Science, 336, 2012
|
|
2M5N
 
 | | Atomic-resolution structure of a cross-beta protofilament | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-{beta} amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6Z3X
 
 | | Crystal structure of the designed antibody DesAb-anti-HSA-P1 | | Descriptor: | CACODYLATE ION, DesAb-anti-HSA-P1, IMIDAZOLE, ... | | Authors: | Costanzi, E, Sormanni, P, Ricagno, S. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based computational design of antibodies targeting structured epitopes.
Sci Adv, 8, 2022
|
|
6DKF
 
 | |
6GRZ
 
 | |
5CKA
 
 | | Human beta-2 microglobulin double mutant W60G-N83V | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
5CKG
 
 | | Human beta-2 microglobulin mutant V85E | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
5CFH
 
 | |
7Z1D
 
 | | Nanobody H11-H6 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H6 nanobody, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1B
 
 | | Nanobody H11-A10 and F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A10, Nanobody F2, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1A
 
 | | Nanobody H11 and F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 Nanobody, H11 Nanobody, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1C
 
 | | Nanobody H11-B5 and H11-F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Nanobody B5, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1E
 
 | | Nanobody H11-H4 Q98R H100E bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H4 Q98R H100E, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z85
 
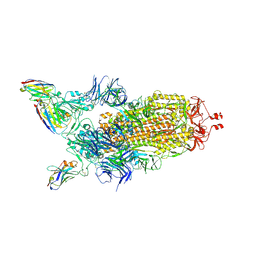 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-B5 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-B5, ... | | Authors: | Weckener, M, Naismith, J.H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9Q
 
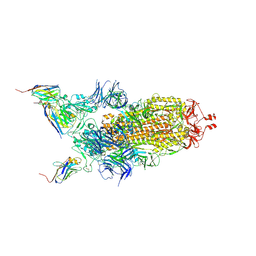 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-A10 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-A10, ... | | Authors: | Weckener, M, Naismith, J.H. | | Deposit date: | 2022-03-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z7X
 
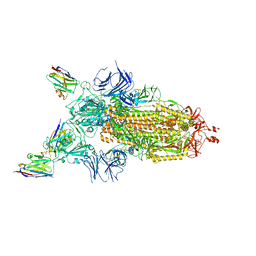 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-H6 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H6, ... | | Authors: | Weckener, M, Naismith, J.H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9R
 
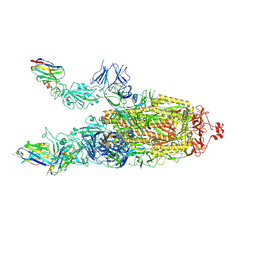 | |
7Z86
 
 | |
7Z6V
 
 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11, ... | | Authors: | Weckener, M, Naismith, J.H, Vogirala, V.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2MVZ
 
 | | Solution Structure for Cyclophilin A from Geobacillus Kaustophilus | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Holliday, M.J, Isern, N.G, Geoffrey, A.S, Zhang, F, Eisenmesser, E.Z. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of GeoCyp: A Thermophilic Cyclophilin with a Novel Substrate Binding Mechanism That Functions Efficiently at Low Temperatures.
Biochemistry, 54, 2015
|
|
