5D5G
 
 | | Structure of colocasia esculenta agglutinin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Biswas, H, Chattopadhyaya, R. | | Deposit date: | 2015-08-10 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure Colocasia esculenta tuber agglutinin at 1.74A resolution and its quaternary interactions
J GLYCOBIO, 6, 2017
|
|
5T85
 
 | |
5T6L
 
 | | Crystal structure of 10E8 Fab in complex with the MPER epitope scaffold T117v2 | | Descriptor: | 10E8 EPITOPE SCAFFOLD T117V2, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Antibody 10E8 FAB HEAVY CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
5T80
 
 | |
5TFW
 
 | | Crystal structure of 10E8 Fab light chain mutant2 against the MPER region of the HIV-1 Env, in complex with T117v2 epitope scaffold | | Descriptor: | 1,2-ETHANEDIOL, 10E8 EPITOPE SCAFFOLD T117V2, Antibody 10E8 FAB HEAVY CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-09-26 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
5Z32
 
 | | LPS bound solution NMR structure of WS2-VR18 | | Descriptor: | VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY-LYS-ASN-LYS-SER-ARG | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
5Z31
 
 | | LPS bound solution structure of WS2-KG18 | | Descriptor: | LYS-ASN-LYS-SER-ARG-VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
5KZC
 
 | | Crystal structure of an HIV-1 gp120 engineered outer domain with a Man9 glycan at position N276, in complex with broadly neutralizing antibody VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Engineered outer domain of gp120, VRC01 Fab heavy chain, ... | | Authors: | Julien, J.-P, Jardine, J.G, Diwanji, D, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-07-24 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
5SY8
 
 | |
5T5B
 
 | |
5T29
 
 | | Crystal structure of 10E8 Fab light chain mutant3, against the MPER region of the HIV-1 Env, in complex with the MPER epitope scaffold T117v2 | | Descriptor: | 10E8 EPITOPE SCAFFOLD T117V2, Antibody 10E8 FAB HEAVY CHAIN, Antibody 10E8 FAB LIGHT CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
3ZW1
 
 | | Structure of Bambl lectine in complex with lewix x antigen | | Descriptor: | BAMBL LECTIN, alpha-D-galactopyranose, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Audfray, A, Claudinon, J, Abounit, S, Ruvoen-Clouet, N, Larson, G, Wimmerova, M, Lependu, J, Romer, W, Varrot, A, Imberty, A. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deciphering the Glycan Preference of Bacterial Lectins by Glycan Array and Molecular Docking with Validation by Microcalorimetry and Crystallography.
Plos One, 8, 2013
|
|
5IDL
 
 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Authors: | Julien, J.P, Ereno-Orbea, J, Jardine, J.G, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen.
Science, 351, 2016
|
|
4XC1
 
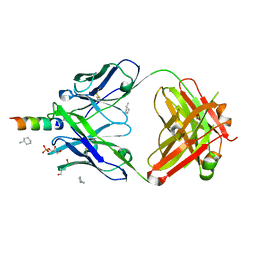 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 GP41: crystals cryoprotected with sn-Glycerol 3-phosphate | | Descriptor: | 4E10 FAB HEAVY CHAIN, 4E10 FAB LIGHT CHAIN, CYCLOHEXYLAMMONIUM ION, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4XCE
 
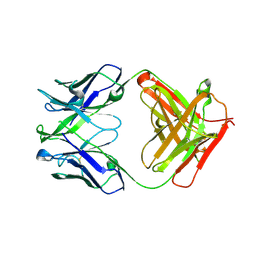 | |
4XBE
 
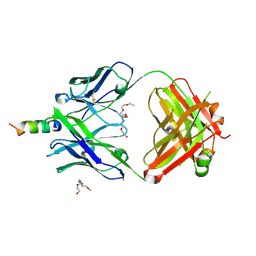 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41: crystals cryoprotected with sphingomyelin (02:0 SM (d18:1/2:0)). | | Descriptor: | 4E10 FAB LIGHT CHAIN, 4E10 Fab heavy chain, PEPTIDE FRAGMENT OF HIV GLYCOPROTEIN (GP41) including the region 671-683 of the MPER, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-16 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4XBP
 
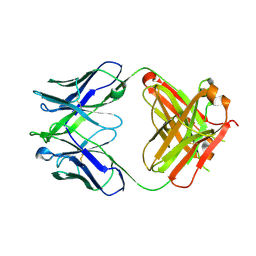 | |
4XCN
 
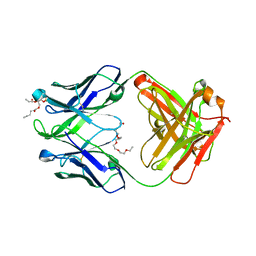 | | Crystal structure of human 4E10 Fab in complex with phosphatidic acid (06:0 PA); 2.9 A resolution | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4XCC
 
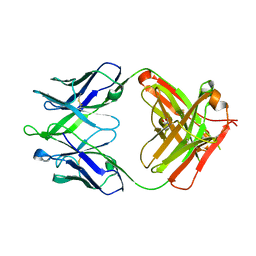 | |
4XCF
 
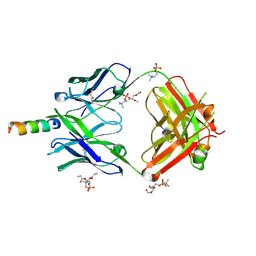 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41; crystals cryoprotected with phosphatidylcholine (03:0 PC) | | Descriptor: | 4E10 Fab heavy chain, 4E10 Fab light chain, MODIFIED FRAGMENT OF HIV-1 GLYCOPROTEIN (GP41) INCLUDING THE MPER REGION 671-683, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4XBG
 
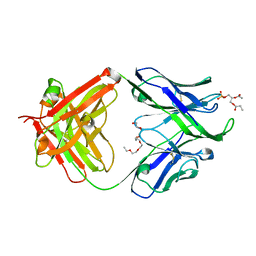 | | Crystal structure of human 4E10 Fab in complex with phosphatidic acid (06:0 PA): 2.73 A resolution | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-16 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4XC3
 
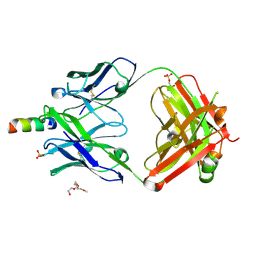 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41; crystals cryoprotected with rac-glycerol 1-phosphate | | Descriptor: | 4E10 Fab heavy chain, 4E10 Fab light chain, MODIFIED FRAGMENT OF HIV-1 GLYCOPROTEIN (GP41) INCLUDING THE MPER REGION 671-683, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4XAW
 
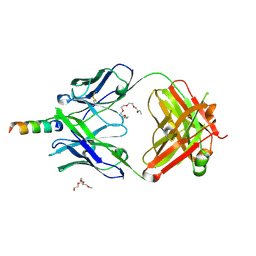 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41: Crystals cryoprotected with phosphatidic acid (08:0 PA) | | Descriptor: | 4E10 Fab heavy chain, 4E10 Fab light chain, FRAGMENT OF HIV GLYCOPROTEIN (GP41) including the MPER region 671-683, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-15 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4XCY
 
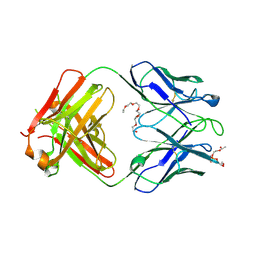 | | Crystal structure of human 4E10 Fab in complex with phosphatidylglycerol (06:0 PG) | | Descriptor: | (2S)-3-{[(R)-{[(2R)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
6NC2
 
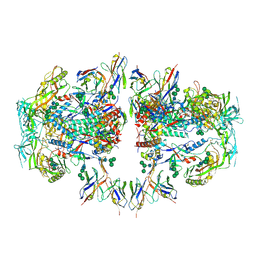 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody ACS202 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Yuan, M, Copps, J, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
