5O9D
 
 | | Crystal structure of R. ruber ADH-A, mutant Y294F, W295A, Y54F, F43H, H39Y | | Descriptor: | (2~{S})-2-methylpentanedioic acid, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer.
FEBS J., 284, 2017
|
|
5O9F
 
 | | Crystal structure of R. ruber ADH-A, mutant Y294F, W295A, Y54F, F43S, H39Y | | Descriptor: | (2~{S})-2-methylpentanedioic acid, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer.
FEBS J., 284, 2017
|
|
5MUB
 
 | | ACC1 Fab fragment in complex with citrullinated C1 epitope of CII (CG05) | | Descriptor: | ACC1 Fab fragment heavy chain, ACC1 Fab fragment light chain, triple-helical peptide containing the citrullinated C1 epitope of collagen type II,Collagen alpha-1(II) chain,triple-helical peptide containing the citrullinated C1 epitope of collagen type II | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MU2
 
 | | ACC1 Fab fragment in complex with CII583-591 (CG10) | | Descriptor: | ACC1 Fab fragment heavy chain, ACC1 Fab fragment light chain, synthetic peptide containing the CII583-591 epitope of collagen type II | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MU0
 
 | | ACC1 Fab fragment in complex with citrullinated C1 epitope of CII (IA03) | | Descriptor: | IA03 peptide containing the citrullinated C1 epitope of collagen type II,Collagen alpha-1(II) chain,IA03 peptide containing the citrullinated C1 epitope of collagen type II, heavy chain of ACC1 antibody Fab fragment, light chain of ACC1 antibody Fab fragment | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-11 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MV3
 
 | | ACC1 Fab fragment in complex with CII583-591 (CG10) | | Descriptor: | heavy chain of ACC1 Fab fragment, light chain of ACC1 Fab fragment, synthetic peptide containing the CII583-591 epitope of collagen type II,Collagen alpha-1(II) chain,synthetic peptide containing the CII583-591 epitope of collagen type II, | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5MV4
 
 | | ACC1 Fab fragment in complex with citrullinated CII616-639 epitope of collagen type II (ptm23) | | Descriptor: | ACC1 antibody Fab fragment, heavy chain, light chain, ... | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
5OD8
 
 | | Crystal structure of the RA-associated mAb B2 (Fab fragment) | | Descriptor: | B2 Fab fragment - Light chain, B2 Fab fragment - heavy chain | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R, Amara, K, Malmstrom, V. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5OD0
 
 | | Crystal structure of ACPA E4 | | Descriptor: | Fab fragment of ACPA E4 - Light chain, Fab fragment of ACPA E4 - heavy chain, GLYCEROL | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5OCY
 
 | |
5OCK
 
 | | Crystal structure of ACPA E4 in complex with CEP1 | | Descriptor: | CEP1 peptide (from enolase), Human ACPA E4 Fab fragment - Heavy chain, Human ACPA E4 Fab fragment - Light chain | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-03 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5ODB
 
 | | Crystal structure of the RA-associated mAb D10 (chimeric Fab fragment) | | Descriptor: | D10 Fab fragment - heavy chain, D10 Fab fragment - light chain, GLYCEROL, ... | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R, Amara, K, Malmstrom, V. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
6QUL
 
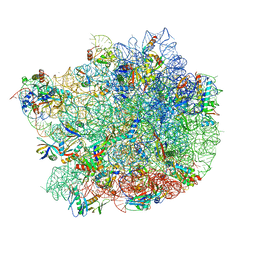 | | Structure of a bacterial 50S ribosomal subunit in complex with the novel quinoxolidinone antibiotic cadazolid | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Scaiola, A, Leibundgut, M, Boehringer, D, Ritz, D. | | Deposit date: | 2019-02-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of translation inhibition by cadazolid, a novel quinoxolidinone antibiotic.
Sci Rep, 9, 2019
|
|
5MMP
 
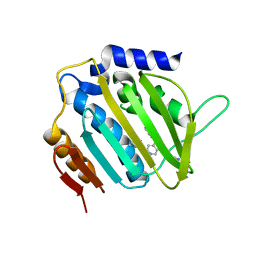 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)-isoquinolin-3-yl]-urea | | Descriptor: | 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)isoquinolin-3-yl]urea, DNA gyrase subunit B | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5MMN
 
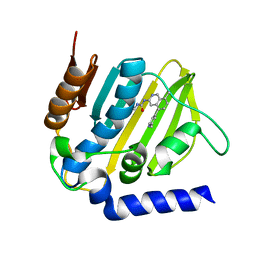 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[8-methyl-5-(2-methyl-pyridin-4-yl)-isoquinolin-3-yl]-urea | | Descriptor: | 1-ethyl-3-[8-methyl-5-(2-methylpyridin-4-yl)isoquinolin-3-yl]urea, DNA gyrase subunit B | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
5MMO
 
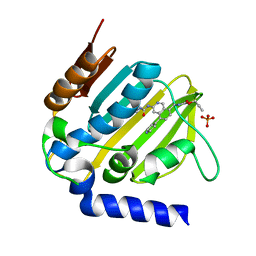 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with [3-(3-ethyl-ureido)-5-(pyridin-4-yl)-isoquinolin-8-yl-methyl]-carbamic acid prop-2-ynyl ester | | Descriptor: | DNA gyrase subunit B, PHOSPHATE ION, prop-2-ynyl ~{N}-[[3-(ethylcarbamoylamino)-5-pyridin-4-yl-isoquinolin-8-yl]methyl]carbamate | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
6I48
 
 | | Structure of P. aeruginosa LpxC with compound 12: (2R)-4-(6-(2-Fluoro-4-methoxyphenyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, 1,2-ETHANEDIOL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6I47
 
 | | Structure of P. aeruginosa LpxC with compound 10: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-1-oxoisoindolin-2-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, (2~{S})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6I49
 
 | | Structure of P. aeruginosa LpxC with compound 17a: (2R)-N-Hydroxy-2-methyl-2-(methylsulfonyl)-4(6((4(morpholinomethyl)phenyl)ethynyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)yl)butanamide | | Descriptor: | (2~{R})-2-methyl-2-methylsulfonyl-4-[6-[2-[4-(morpholin-4-ylmethyl)phenyl]ethynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6I4A
 
 | | Structure of P. aeruginosa LpxC with compound 18d: (2R)-N-Hydroxy-4-(6-((1-(hydroxymethyl)cyclopropyl)buta-1,3-diyn-1-yl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-[4-[1-(hydroxymethyl)cyclopropyl]buta-1,3-diynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6I46
 
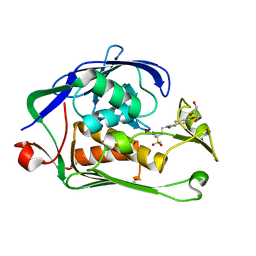 | | Structure of P. aeruginosa LpxC with compound 8: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-2-oxooxazol-3(2H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[5-(2-fluoranyl-4-methoxy-phenyl)-2-oxidanylidene-1,3-oxazol-3-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, GLYCEROL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
1QPB
 
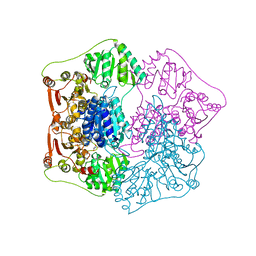 | | PYRUVATE DECARBOYXLASE FROM YEAST (FORM B) COMPLEXED WITH PYRUVAMIDE | | Descriptor: | MAGNESIUM ION, PYRUVAMIDE, PYRUVATE DECARBOXYLASE (FORM B), ... | | Authors: | Lu, G, Dobritzsch, D, Schneider, G. | | Deposit date: | 1999-11-26 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Basis of Substrate Activation in Yeast Pyruvate Decarboxylase a Crystallographic and Kinetic Study
Eur.J.Biochem., 267, 2000
|
|
2QA1
 
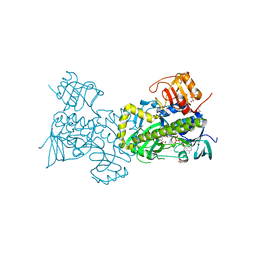 | | Crystal structure of PgaE, an aromatic hydroxylase involved in angucycline biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
2QA2
 
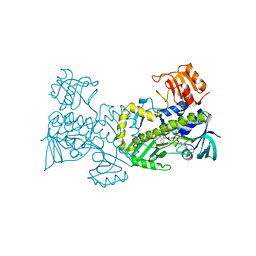 | | Crystal structure of CabE, an aromatic hydroxylase from angucycline biosynthesis, determined to 2.7 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyketide oxygenase CabE | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
7QLB
 
 | | SMYD3 in complex with fragment FL06268 | | Descriptor: | 1-methylimidazole-4-sulfonamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2021-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening
Rsc Med Chem, 2024
|
|
