6QBT
 
 | |
6QBV
 
 | |
6QBW
 
 | |
6HV9
 
 | | S. cerevisiae CMG-Pol epsilon-DNA | | Descriptor: | Cell division control protein 45, DNA (5'-D(*GP*CP*AP*GP*CP*CP*AP*CP*GP*CP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*GP*CP*GP*TP*GP*GP*CP*TP*GP*C)-3'), ... | | Authors: | Abid Ali, F, Purkiss, A.G, Cheung, A, Costa, A. | | Deposit date: | 2018-10-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome.
Nat Commun, 9, 2018
|
|
6HV8
 
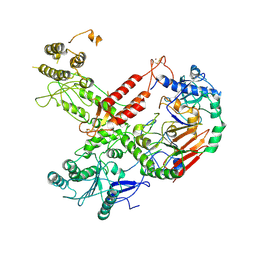 | | Cryo-EM structure of S. cerevisiae Polymerase epsilon deltacat mutant | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit B, ZINC ION | | Authors: | Goswami, P, Purkiss, A, Cheung, A, Costa, A. | | Deposit date: | 2018-10-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome.
Nat Commun, 9, 2018
|
|
7P30
 
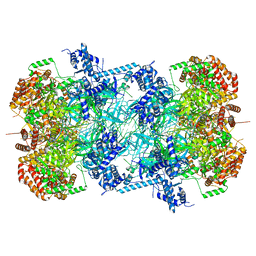 | | 3.0 A resolution structure of a DNA-loaded MCM double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (53-MER), ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-02-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P5Z
 
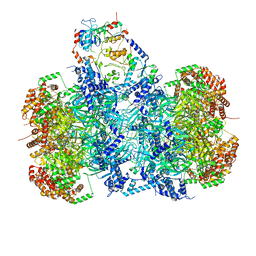 | | Structure of a DNA-loaded MCM double hexamer engaged with the Dbf4-dependent kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 7, ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-02-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7B62
 
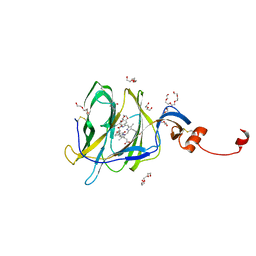 | | Crystal structure of SARS-CoV-2 spike protein N-terminal domain in complex with biliverdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pye, V.E, Rosa, A, Roustan, C, Cherepanov, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7AML
 
 | | RET/GDNF/GFRa1 extracellular complex Cryo-EM structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-10-09 | | Release date: | 2021-01-13 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
7AMK
 
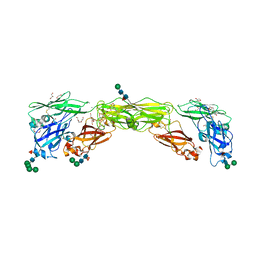 | | Zebrafish RET Cadherin Like Domains 1 to 4. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Purkiss, A.G, McDonald, N.Q, Goodman, K.M, Narowtek, A, Knowles, P.P. | | Deposit date: | 2020-10-09 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
7QHS
 
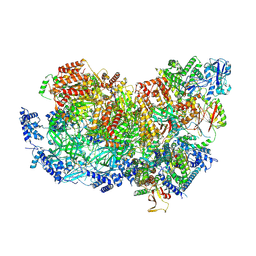 | | S. cerevisiae CMGE nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
7AB8
 
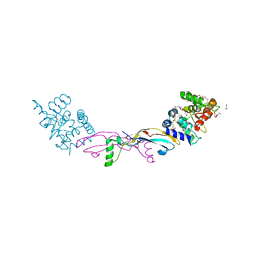 | | Crystal structure of a GDNF-GFRalpha1 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
7Z1Z
 
 | | MVV strand transfer complex (STC) intasome in complex with LEDGF/p75 at 3.5 A resolution | | Descriptor: | DNA (37-MER), DNA (5'-D(*GP*CP*TP*GP*CP*GP*AP*GP*AP*TP*CP*CP*GP*CP*TP*CP*CP*GP*GP*TP*G)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*TP*AP*GP*GP*GP*TP*G)-3'), ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Cherepanov, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multivalent interactions essential for lentiviral integrase function.
Nat Commun, 13, 2022
|
|
7Z13
 
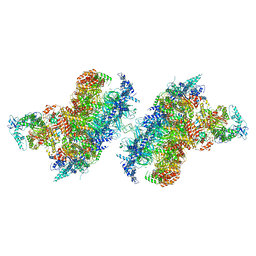 | | S. cerevisiae CMGE dimer nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
6HJR
 
 | |
6HJN
 
 | | Structure of Influenza Hemagglutinin ectodomain (A/duck/Alberta/35/76) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Benton, D.J, Rosenthal, P.B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Influenza hemagglutinin membrane anchor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HKG
 
 | | Structure of FISW84 Fab Fragment | | Descriptor: | FISW84 Fab Fragment Heavy Chain, FISW84 Fab Fragment Light Chain, SULFATE ION | | Authors: | Benton, D.J, Rosenthal, P.B. | | Deposit date: | 2018-09-06 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Influenza hemagglutinin membrane anchor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HJQ
 
 | |
6HJP
 
 | |
6RNY
 
 | | PFV intasome - nucleosome strand transfer complex | | Descriptor: | DNA (108-MER), DNA (128-MER), DNA (33-MER), ... | | Authors: | Pye, V.E, Renault, L, Maskell, D.P, Cherepanov, P, Costa, A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
6F07
 
 | | CBF3 Core Complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Suppressor of kinetochore protein 1, ZINC ION | | Authors: | Leber, V, Singleton, M.R. | | Deposit date: | 2017-11-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for assembly of the CBF3 kinetochore complex.
EMBO J., 37, 2018
|
|
8QXO
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State V - Depleted relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXN
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State IV - Depleted relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXL
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State II - Hemi-relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXK
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State I - Tense | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
