6DDQ
 
 | | Crystal structure of the double mutant (R39Q/D52N) of the full-length NT5C2 in the basal state | | Descriptor: | 1,2-ETHANEDIOL, Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DE2
 
 | | Crystal structure of the double mutant (D52N/L375F) of the full-length NT5C2 in the active state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDZ
 
 | | Crystal structure of the double mutant (D52N/R238W) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDO
 
 | | Crystal structure of the single mutant (D52N) of the full-length NT5C2 in the basal state | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
7S22
 
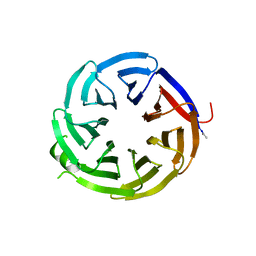 | | Crystal structure of alpha-COP-WD40 domain | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S23
 
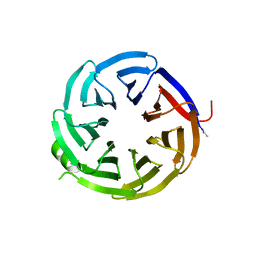 | | Crystal structure of alpha-COP-WD40 domain, Y139A mutant | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S16
 
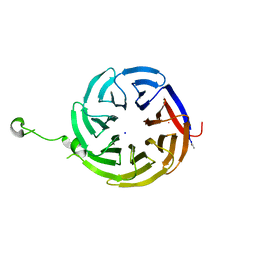 | | Crystal structure of alpha-COP-WD40 domain R57A mutant | | Descriptor: | Coatomer subunit alpha, SODIUM ION | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
5JD3
 
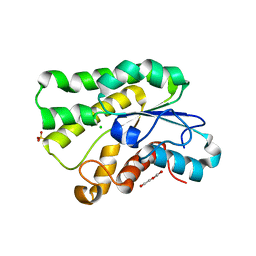 | | Crystal structure of LAE5, an alpha/beta hydrolase enzyme from the metagenome of Lake Arreo, Spain | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, LAE5, ... | | Authors: | Stogios, P.J, Xu, X, Nocek, B, Cui, H, Yim, V, Martinez-Martinez, M, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To Be Published
|
|
5JD4
 
 | | Crystal structure of LAE6 Ser161Ala mutant, an alpha/beta hydrolase enzyme from the metagenome of Lake Arreo, Spain | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Xu, X, Alcaide, M, Yim, V, Cui, H, Martinez-Martinez, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of LAE6 Ser161Ala mutant, an alpha/beta hydrolase enzyme from the metagenome of Lake Arreo, Spain
To Be Published
|
|
5JD6
 
 | | Crystal structure of MGS-MChE2, an alpha/beta hydrolase enzyme from the metagenome of sediments from the lagoon of Mar Chica, Morocco | | Descriptor: | MGS-MChE2 | | Authors: | Stogios, P.J, Xu, X, Nocek, B, Yim, V, Cui, H, Martinez-Martinez, M, Golyshin, P.N, Yakima, M.M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Crystal structure of MGS-MChE2, an alpha/beta hydrolase enzyme from the metagenome of sediments from the lagoon of Mar Chica, Morocco
To Be Published
|
|
5JD5
 
 | | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy | | Descriptor: | CHLORIDE ION, MGS-MilE3 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Martinez-Martinez, M, Chernikova, T.N, Golyshin, P.N, Yakimov, M.M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy
To Be Published
|
|
3EY6
 
 | | Crystal structure of the FK506-binding domain of human FKBP38 | | Descriptor: | FK506-binding protein 8 | | Authors: | Parthier, C, Maestre-Martinez, M, Neumann, P, Edlich, F, Fischer, G, Luecke, C, Stubbs, M.T. | | Deposit date: | 2008-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A charge-sensitive loop in the FKBP38 catalytic domain modulates Bcl-2 binding.
J.Mol.Recognit., 24, 2011
|
|
5OHF
 
 | | Globin sensor domain of AfGcHK (FeIII form) in complex with cyanide, partially reduced | | Descriptor: | CHLORIDE ION, CYANIDE ION, Globin-coupled histidine kinase, ... | | Authors: | Skalova, T, Kolenko, P, Dohnalek, J, Stranava, M, Martinkova, M. | | Deposit date: | 2017-07-16 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coordination and redox state-dependent structural changes of the heme-based oxygen sensor AfGcHK associated with intraprotein signal transduction.
J. Biol. Chem., 292, 2017
|
|
5OHE
 
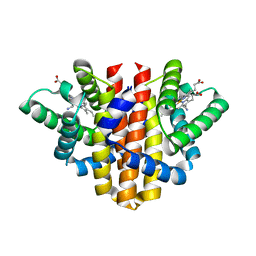 | | Globin sensor domain of AfGcHK (FeIII form) in complex with cyanide | | Descriptor: | CHLORIDE ION, CYANIDE ION, Globin-coupled histidine kinase, ... | | Authors: | Skalova, T, Kolenko, P, Dohnalek, J, Stranava, M, Martinkova, M. | | Deposit date: | 2017-07-16 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Coordination and redox state-dependent structural changes of the heme-based oxygen sensor AfGcHK associated with intraprotein signal transduction.
J. Biol. Chem., 292, 2017
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
6OTD
 
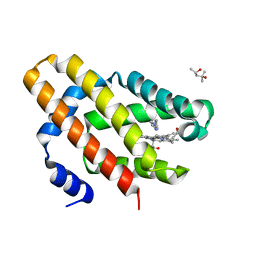 | | Globin sensor domain of AfGcHK in monomeric form, with imidazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Globin-coupled histidine kinase, IMIDAZOLE, ... | | Authors: | Skalova, T, Dohnalek, J, Kolenko, P, Stranava, M, Lengalova, A, Martinkova, M. | | Deposit date: | 2019-05-03 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Disruption of the dimerization interface of the sensing domain in the dimeric heme-based oxygen sensorAfGcHK abolishes bacterial signal transduction.
J.Biol.Chem., 295, 2020
|
|
2F2D
 
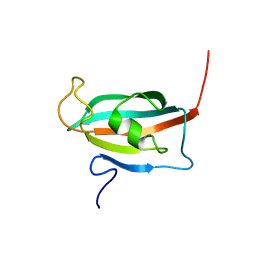 | | Solution structure of the FK506-binding domain of human FKBP38 | | Descriptor: | 38 kDa FK-506 binding protein homolog, FKBP38 | | Authors: | Maestre-Martinez, M, Edlich, F, Jarczowski, F, Weiwad, M, Fischer, G, Luecke, C. | | Deposit date: | 2005-11-16 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the FK506-Binding Domain of Human FKBP38
J.BIOMOL.NMR, 34, 2006
|
|
8QKE
 
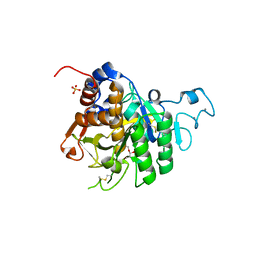 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (MH-13) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Peptidomimetic Inhibitor (MH-13), ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Insights from structure-activity relationships and the binding mode of peptidic alpha-ketoamide inhibitors of the malaria drug target subtilisin-like SUB1.
Eur.J.Med.Chem., 269, 2024
|
|
8QKG
 
 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (MAM-125) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Peptidomimetic Inhibitor (MAM-125), ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Insights from structure-activity relationships and the binding mode of peptidic alpha-ketoamide inhibitors of the malaria drug target subtilisin-like SUB1.
Eur.J.Med.Chem., 269, 2024
|
|
8QKJ
 
 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (MAM-133) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Peptidomimetic Inhibitor (MAM-133), ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Insights from structure-activity relationships and the binding mode of peptidic alpha-ketoamide inhibitors of the malaria drug target subtilisin-like SUB1.
Eur.J.Med.Chem., 269, 2024
|
|
1FG4
 
 | | STRUCTURE OF TRYPAREDOXIN II | | Descriptor: | TRYPAREDOXIN II | | Authors: | Hofmann, B, Budde, H, Bruns, K, Guerrero, S.A, Kalisz, H.M, Menge, U, Montemartini, M, Nogoceke, E, Steinert, P, Wissing, J.B, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-07-28 | | Release date: | 2001-04-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
1BW0
 
 | | CRYSTAL STRUCTURE OF TYROSINE AMINOTRANSFERASE FROM TRYPANOSOMA CRUZI | | Descriptor: | PROTEIN (TYROSINE AMINOTRANSFERASE) | | Authors: | Blankenfeldt, W, Montemartini, M, Hunter, G.R, Kalisz, H.M, Nowicki, C, Hecht, H.J. | | Deposit date: | 1998-09-28 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi tyrosine aminotransferase: substrate specificity is influenced by cofactor binding mode.
Protein Sci., 8, 1999
|
|
2KDU
 
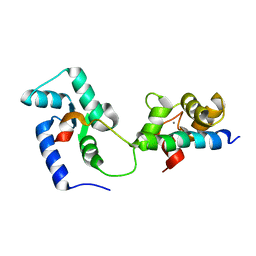 | | Structural basis of the Munc13-1/Ca2+-Calmodulin interaction: A novel 1-26 calmodulin binding motif with a bipartite binding mode | | Descriptor: | CALCIUM ION, Calmodulin, Protein unc-13 homolog A | | Authors: | Rodriguez-Castaneda, F.A, Maestre-Martinez, M, Coudevylle, N, Dimova, K, Jahn, O, Junge, H, Becker, S, Brose, N, Carlomagno, T, Griesinger, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity.
Embo J., 29, 2010
|
|
2N6J
 
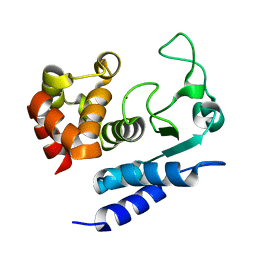 | | Solution structure of Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile | | Descriptor: | ZINC ION, Zinc metalloprotease Zmp1 | | Authors: | Banci, L, Cantini, F, Scarselli, M, Rubino, J.T, Martinelli, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of zinc-bound Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile.
J.Biol.Inorg.Chem., 21, 2016
|
|
