5CCH
 
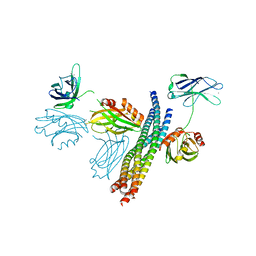 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (short unit cell form) | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Zeldin, O.B, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
5CCI
 
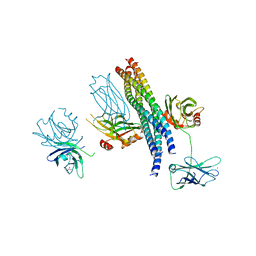 | | Structure of the Mg2+-bound synaptotagmin-1 SNARE complex (short unit cell form) | | Descriptor: | MAGNESIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
5CCG
 
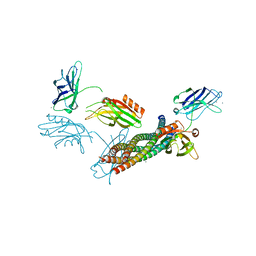 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Zhou, Q, Zhao, M, Lyubimov, A.Y, Uervirojnangkoorn, M, Zeldin, O.B, Weis, W.I, Brunger, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis.
Nature, 525, 2015
|
|
5TOO
 
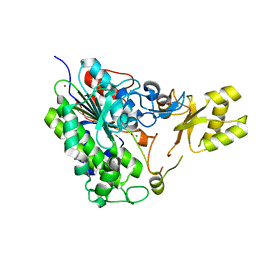 | | Crystal structure of alkaline phosphatase PafA T79S, N100A, K162A, R164A mutant | | Descriptor: | Alkaline phosphatase PafA, CHLORIDE ION, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, AlSadhan, I, Herschlag, D. | | Deposit date: | 2016-10-18 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
1N4V
 
 | |
1N4U
 
 | |
1N4W
 
 | |
4FTH
 
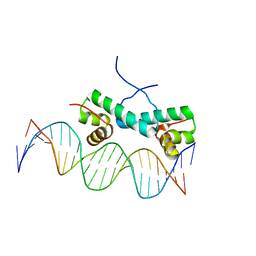 | | Crystal Structure of NtrC4 DNA-binding domain bound to double-stranded DNA | | Descriptor: | 5'-D(*AP*CP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*GP*CP*AP*T)-3', 5'-D(P*GP*AP*TP*GP*CP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*A)-3', Transcriptional regulator (NtrC family) | | Authors: | Vidangos, N.K, Heideker, J, Lyubimov, A.Y, Lamers, M, Huo, Y, Pelton, J.G, Ton, J, Gralla, J.D, Kuriyan, J, Berger, J.M, Wemmer, D.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | DNA Recognition by a sigma (54) Transcriptional Activator from Aquifex aeolicus.
J.Mol.Biol., 426, 2014
|
|
6APF
 
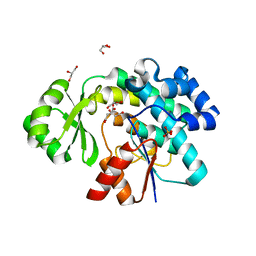 | | Trans-acting transferase from Disorazole synthase complexed with Citrate. | | Descriptor: | CITRIC ACID, DisD protein, GLYCEROL, ... | | Authors: | Mathews, I.I, Lyubimov, A, Soltis, M, Khosla, C, Cohen, A, Robbins, T. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
4PNJ
 
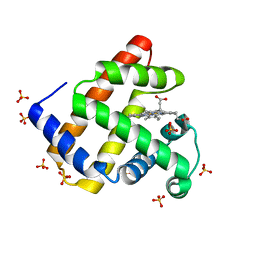 | | Recombinant Sperm Whale P6 Myoglobin Solved with Single Pulse Free Electron Laser Data | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Cohen, A, Gonzalez, A, Lam, W, Lyubimov, A, Sauter, N, Tsai, Y, Uervirojnangkoorn, M, Brunger, A, Soltis, M. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Goniometer-based femtosecond crystallography with X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5TPQ
 
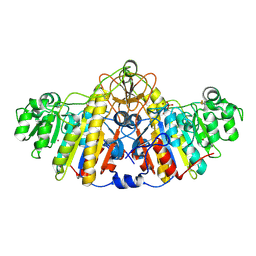 | | E. coli alkaline phosphatase D101A, D153A, R166S, E322A, K328A mutant | | Descriptor: | Alkaline phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Sunden, F, AlSadhan, I, Lyubimov, A.Y, Doukov, T, Swan, J, Herschlag, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
6CHB
 
 | |
6CH9
 
 | |
6CH7
 
 | |
6CH8
 
 | |
6N6P
 
 | | Crystal structure of [FeFe]-hydrogenase in the presence of 7 mM Sodium dithionite | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Peters, J.W. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
6N59
 
 | | 1.0 Angstrom crystal structure of [FeFe]-hydrogenase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Artz, J.H, Peters, J.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
5CCJ
 
 | |
1MXT
 
 | | Atomic resolution structure of Cholesterol oxidase (Streptomyces sp. SA-COO) | | Descriptor: | CHOLESTEROL OXIDASE, FLAVIN-N7 PROTONATED-ADENINE DINUCLEOTIDE, OXYGEN MOLECULE, ... | | Authors: | Vrielink, A, Lario, P.I. | | Deposit date: | 2002-10-03 | | Release date: | 2003-02-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Sub-atomic resolution crystal structure of cholesterol oxidase:
What atomic resolution crystallography reveals about enzyme mechanism and the role of FAD cofactor in redox activity
J.Mol.Biol., 326, 2003
|
|
