1HGD
 
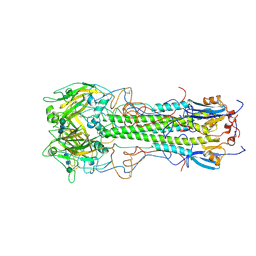 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGI
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGF
 
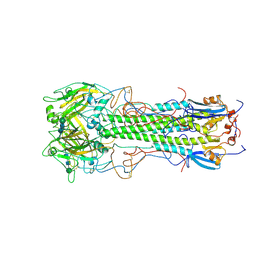 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
2PRO
 
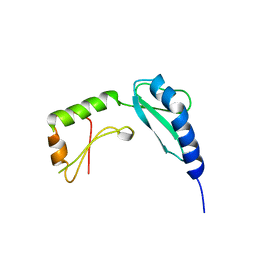 | |
1HGE
 
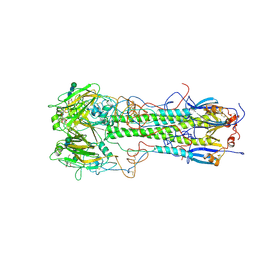 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGH
 
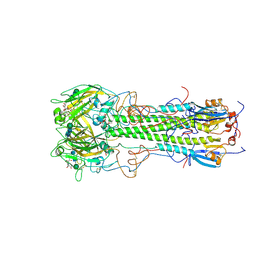 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGJ
 
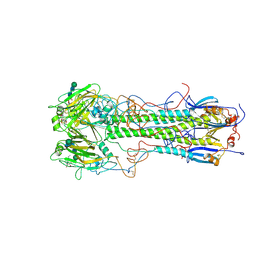 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGG
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
3PRO
 
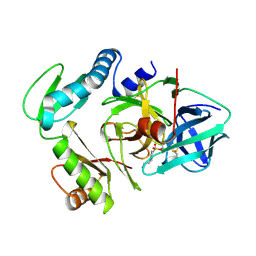 | | ALPHA-LYTIC PROTEASE COMPLEXED WITH C-TERMINAL TRUNCATED PRO REGION | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ALPHA-LYTIC PROTEASE | | Authors: | Sauter, N.K, Mau, T, Rader, S.D, Agard, D.A. | | Deposit date: | 1998-08-26 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of alpha-lytic protease complexed with its pro region.
Nat.Struct.Biol., 5, 1998
|
|
4PRO
 
 | |
4WMG
 
 | | Structure of hen egg-white lysozyme from a microfludic harvesting device using synchrotron radiation (2.5A) | | Descriptor: | Lysozyme C | | Authors: | Lyubimov, A.Y, Murray, T.D, Koehl, A, Uervirojnangkoorn, M, Zeldin, O.B, Cohen, A.E, Soltis, S.M, Baxter, E.M, Brewster, A.S, Sauter, N.K, Brunger, A.T, Berger, J.M. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4FBY
 
 | | fs X-ray diffraction of Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Hellmich, J, Tran, R, Hattne, J, Laksmono, H, Gloeckner, C, Echols, N, Sierra, R.G, Sellberg, J, Lassalle-Kaiser, B, Gildea, R.J, Glatzel, P, Grosse-Kunstleve, R.W, Latimer, M.J, Mcqueen, T.A, Difiore, D, Fry, A.R, Messerschmidt, M.M, Miahnahri, A, Schafer, D.W, Seibert, M.M, Sokaras, D, Weng, T.-C, Zwart, P.H, White, W.E, Adams, P.D, Bogan, M.J, Boutet, S, Williams, G.J, Messinger, J, Sauter, N.K, Zouni, A, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (6.56 Å) | | Cite: | Room temperature femtosecond X-ray diffraction of photosystem II microcrystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4IXR
 
 | | RT fs X-ray diffraction of Photosystem II, first illuminated state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
4IXQ
 
 | | RT fs X-ray diffraction of Photosystem II, dark state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.7 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
4WG1
 
 | | Room temperature crystal structure of lysozyme determined by serial synchrotron crystallography (micro focused beam - crystFEL) | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Brewster, A.S, Kapp, U, Shilova, A, Weimhausen, B, Sauter, N.K, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-09-17 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TNJ
 
 | | RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 4.5 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNI
 
 | | RT XFEL structure of Photosystem II 500 ms after the third illumination at 4.6 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNK
 
 | | RT XFEL structure of Photosystem II 250 microsec after the third illumination at 5.2 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNH
 
 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.900007 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNL
 
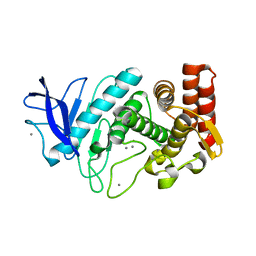 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4OW3
 
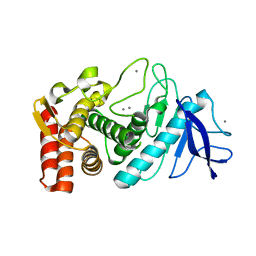 | | Thermolysin structure determined by free-electron laser | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Hattne, J, Echols, N, Tran, R, Kern, J, Gildea, R.J, Brewster, A.S, Alonso-Mori, R, Glockner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, White, W.E, Schafer, D.W, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Glatzel, P, Zwart, P.H, Grosse-Kunstleve, R.W, Bogan, M.J, Messerschmidt, M, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Yano, J, Bergmann, U, Yachandra, V.K, Adams, P.D, Sauter, N.K. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers.
Nat.Methods, 11, 2014
|
|
5KJ7
 
 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) - from XFEL diffraction | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Lyubimov, A.Y, Uervirojnangkoorn, M, Zhou, Q, Zhao, M, Sauter, N.K, Brewster, A.S, Weis, W.I, Brunger, A.T. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Advances in X-ray free electron laser (XFEL) diffraction data processing applied to the crystal structure of the synaptotagmin-1 / SNARE complex.
Elife, 5, 2016
|
|
4QX0
 
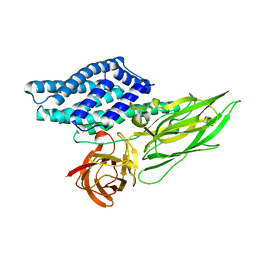 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
