5I7L
 
 | |
8WA4
 
 | |
8KH6
 
 | | Crystal structure of FGFR4 kinase domain with 8r | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[5-bromanyl-6-(hydroxymethyl)-3-methoxy-pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH8
 
 | | Crystal structure of FGFR4(V550L) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH9
 
 | | Crystal structure of FGFR4(V550M) kinase domain with 8z | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8KH7
 
 | | Crystal structure of FGFR4 kinase domain with 8zc | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-3-[6-methanoyl-5-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3-(2-morpholin-4-ylethoxy)pyridin-2-yl]urea, Fibroblast growth factor receptor 4, ... | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8SU9
 
 | | E. coli SIR2-HerA complex (hexamer HerA bound with dodecamer Sir2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside triphosphate hydrolase, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-05-11 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8SUW
 
 | | E. coli SIR2-HerA complex (dodecamer SIR2 bound 4 protomers of HerA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside triphosphate hydrolase, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-05-13 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
3SQJ
 
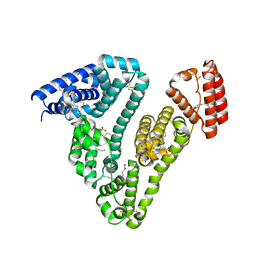 | | Recombinant human serum albumin from transgenic plant | | Descriptor: | MYRISTIC ACID, Serum albumin | | Authors: | He, Y, Yang, D. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Large-scale production of functional human serum albumin from transgenic rice seeds.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6JKV
 
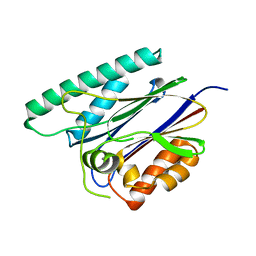 | | PppA, a key regulatory component of T6SS in Pseudomonas aeruginosa | | Descriptor: | MANGANESE (II) ION, PppA | | Authors: | Wang, T, Liu, L, Wu, Y, Li, D. | | Deposit date: | 2019-03-02 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PppA from Pseudomonas aeruginosa, a key regulatory component of type VI secretion systems.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
8SXX
 
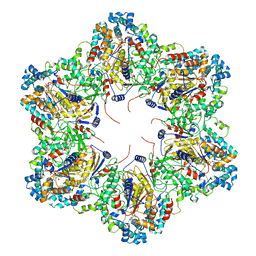 | | E. coli dodecamer SIR2 | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8SUB
 
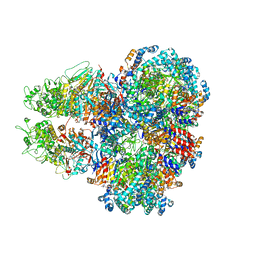 | | E. coli SIR2-HerA complex (dodecamer SIR2 pentamer HerA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside triphosphate hydrolase, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-05-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8UAE
 
 | | E. coli Sir2_HerA complex (12:6) with ATPgamaS | | Descriptor: | MAGNESIUM ION, Nucleoside triphosphate hydrolase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8UAF
 
 | | E. coli Sir2_HerA complex (12:6) bound with NAD+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
7Y4T
 
 | | Crystal structure of cMET kinase domain bound by compound 9I | | Descriptor: | 2-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-(3-nitrophenyl)pyridazin-3-one, Hepatocyte growth factor receptor | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7Y4U
 
 | | Crystal structure of cMET kinase domain bound by compound 9Y | | Descriptor: | Hepatocyte growth factor receptor, ~{N}-methyl-4-[1-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-oxidanylidene-pyridazin-3-yl]-2-(trifluoromethyl)benzamide | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7WCX
 
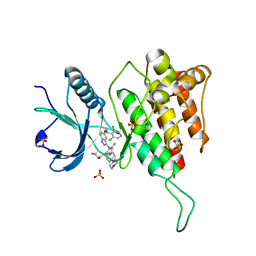 | | Crystal structure of FGFR4(V550M) kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[2-[[5-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1~{H}-indazol-3-yl]amino]-3-fluoranyl-5-(4-morpholin-4-ylpiperidin-1-yl)phenyl]propanamide | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7WCT
 
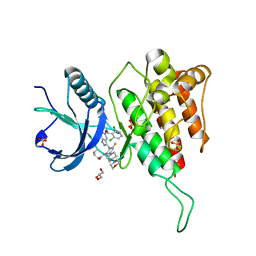 | | Crystal structure of FGFR4 kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7WCW
 
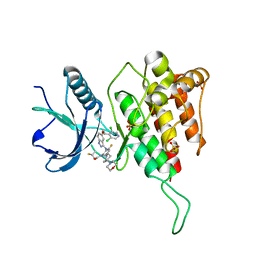 | | Crystal structure of FGFR4(V550L) kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[2-[[5-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1~{H}-indazol-3-yl]amino]-3-fluoranyl-5-(4-morpholin-4-ylpiperidin-1-yl)phenyl]propanamide | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.317 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
8TJY
 
 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TK1
 
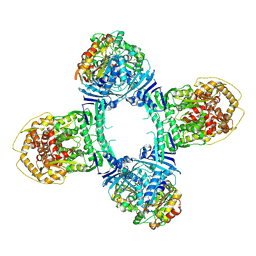 | | Structure of Gabija AB complex 1 | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TK0
 
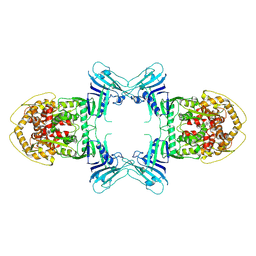 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7TV6
 
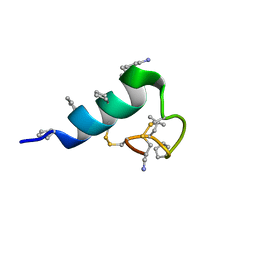 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: native loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV5
 
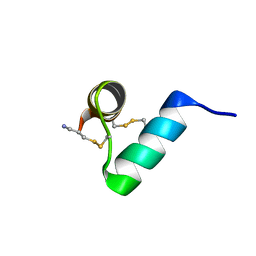 | | Disulfide-rich venom peptide lasiocepsin: P20A mutant | | Descriptor: | Lasiocepsin | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV8
 
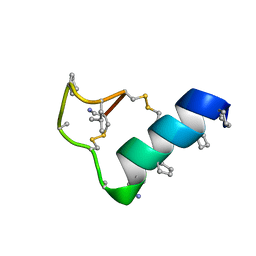 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: D-Ala modified loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
