1QK7
 
 | |
3HW5
 
 | | crystal structure of avian influenza virus PA_N in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Polymerase acidic protein | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Liang, S, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
1NIX
 
 | |
1QFD
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-AMYLASE INHIBITOR (AAI) | | Descriptor: | PROTEIN (ALPHA-AMYLASE INHIBITOR) | | Authors: | Lu, S, Deng, P, Liu, X, Luo, J, Han, R, Gu, X, Liang, S, Wang, X, Feng, L, Lozanov, V, Patthy, A, Pongor, S. | | Deposit date: | 1999-04-08 | | Release date: | 1999-07-16 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major alpha-amylase inhibitor of the crop plant amaranth.
J.Biol.Chem., 274, 1999
|
|
4CC0
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN JAGGED-1, ... | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4CC1
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4CBZ
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN JAGGED-1, alpha-L-fucopyranose | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
2JTB
 
 | |
1Y29
 
 | |
4FO2
 
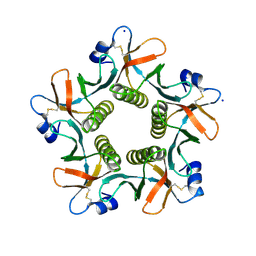 | | Heat-labile enterotoxin LT-IIb-B5(T13I) mutant | | Descriptor: | ACETATE ION, Heat-labile enterotoxin IIB, B chain, ... | | Authors: | Cody, V, Pace, J, Nawar, H, Liang, S, Connell, T, Hajishengallis, G. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-activity correlations of variant forms of the B pentamer of Escherichia coli type II heat-labile enterotoxin LT-IIb with Toll-like receptor 2 binding.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FP5
 
 | | Heat-labile enterotoxin ILT-IIbB5 S74A mutant | | Descriptor: | Heat-labile enterotoxin IIB, B chain, SULFATE ION | | Authors: | Cody, V, Pace, J, Nawar, H, Liang, S, Connell, T, Hajishengallis, G. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity correlations of variant forms of the B pentamer of Escherichia coli type II heat-labile enterotoxin LT-IIb with Toll-like receptor 2 binding.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8KEI
 
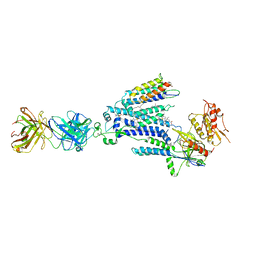 | | Cryo-EM structure of NADPH oxidase 2 in complex with p22phox and EROS | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liang, S.Y, Liu, A.J, Liu, Y.Z, Ye, R.D. | | Deposit date: | 2023-08-11 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for EROS binding to human phagocyte NADPH oxidase NOX2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SKL
 
 | | PTP1B in complex with 182 | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-fluoro-3-hydroxy-7-(3-hydroxy-3-methylbutoxy)naphthalen-2-yl]-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, CHLORIDE ION, ... | | Authors: | Kershaw, N.J, Babon, J.J, Chen, H, Tiganis, T. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A small molecule inhibitor of PTP1B and PTPN2 enhances T cell anti-tumor immunity.
Nat Commun, 14, 2023
|
|
6ZFP
 
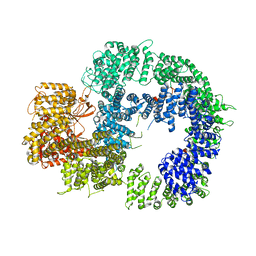 | | Cryo-EM structure of DNA-PKcs (State 2) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHE
 
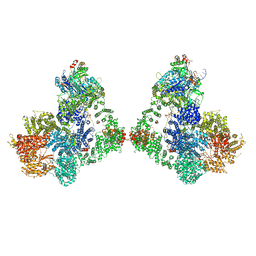 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
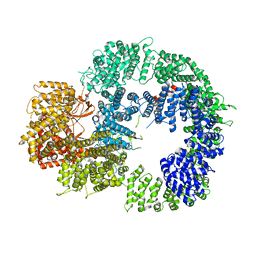 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
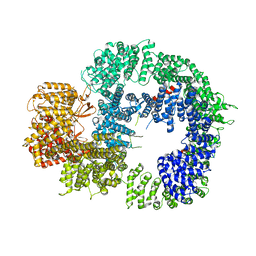 | | Cryo-EM structure of DNA-PKcs (State 3) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
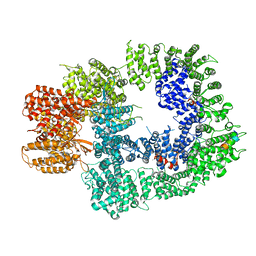 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH8
 
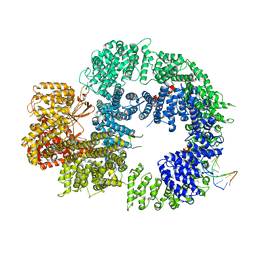 | | Cryo-EM structure of DNA-PKcs:DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*T)-3'), DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHA
 
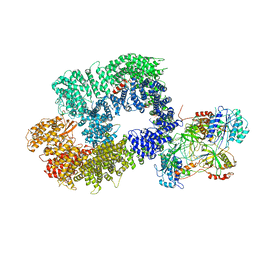 | | Cryo-EM structure of DNA-PK monomer | | Descriptor: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3W2D
 
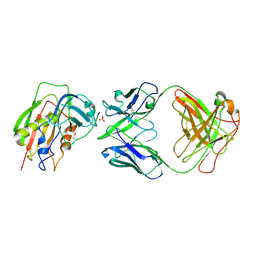 | | Crystal Structure of Staphylococcal Eenterotoxin B in complex with a novel neutralization monoclonal antibody Fab fragment | | Descriptor: | Enterotoxin type B, Monoclonal Antibody 3E2 Fab figment heavy chain, Monoclonal Antibody 3E2 Fab figment light chain, ... | | Authors: | Liang, S.Y, Hu, S, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the neutralization and specificity of Staphylococcal enterotoxin B against its MHC Class II binding site.
MAbs, 6, 2014
|
|
4Z69
 
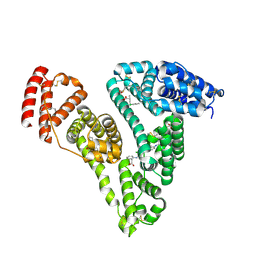 | | Human serum albumin complexed with palmitic acid and diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, PALMITIC ACID, PENTADECANOIC ACID, ... | | Authors: | Zhang, Y, Yang, F. | | Deposit date: | 2015-04-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural basis of non-steroidal anti-inflammatory drug diclofenac binding to human serum albumin.
Chem.Biol.Drug Des., 86, 2015
|
|
7JU8
 
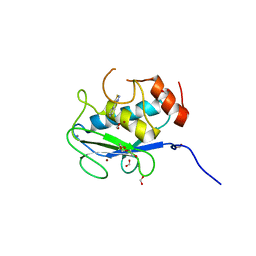 | | X-ray structure of MMP-13 in Complex with 4-(1,2,3-thiadiazol-4-yl)pyridine | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)pyridine, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indole Inhibitors of MMP-13 for Arthritic Disorders
Acs Omega, 6, 2021
|
|
5TTS
 
 | | Jak3 with covalent inhibitor 4 | | Descriptor: | 1-{(3R)-3-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}propan-1-one, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
5TTU
 
 | | Jak3 with covalent inhibitor 7 | | Descriptor: | 1-[(3aR,7aR)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydro-6H-pyrrolo[2,3-c]pyridin-6-yl]propan-1-one, SULFATE ION, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
