2GSG
 
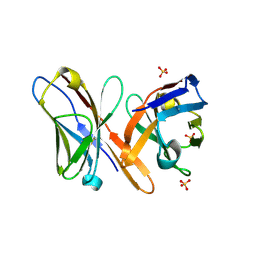 | | Crystal structure of the Fv fragment of a monoclonal antibody specific for poly-glutamine | | Descriptor: | SULFATE ION, monoclonal antibody heavy chain, monoclonal antibody light chain | | Authors: | Li, P, Huey-Tubman, K.E, West Jr, A.P, Bennett, M.J, Bjorkman, P.J. | | Deposit date: | 2006-04-26 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4PP8
 
 | |
8Z1V
 
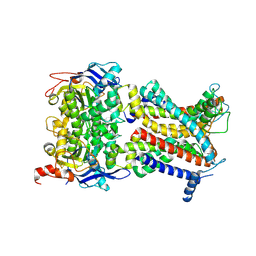 | | Cryo-EM structure of Escherichia coli DppBCDF in the resting state | | Descriptor: | Dipeptide transport ATP-binding protein DppD, Dipeptide transport ATP-binding protein DppF, Dipeptide transport system permease protein DppB, ... | | Authors: | Li, P, Huang, Y. | | Deposit date: | 2024-04-12 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural characterization of the ABC transporter DppABCDF in Escherichia coli reveals insights into dipeptide acquisition.
Plos Biol., 23, 2025
|
|
8Z1Y
 
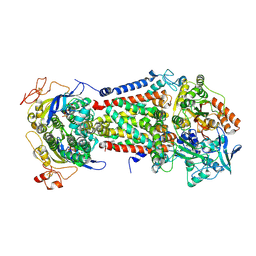 | | Cryo-EM structure of Escherichia coli DppABCDF in the pre-catalytic state | | Descriptor: | Dipeptide transport ATP-binding protein DppD, Dipeptide transport ATP-binding protein DppF, Dipeptide transport system permease protein DppB, ... | | Authors: | Li, P, Huang, Y. | | Deposit date: | 2024-04-12 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural characterization of the ABC transporter DppABCDF in Escherichia coli reveals insights into dipeptide acquisition.
Plos Biol., 23, 2025
|
|
8Z1Z
 
 | |
8Z21
 
 | |
8Z1W
 
 | | Cryo-EM structure of Escherichia coli DppBCDF complex bound to ATPgammaS | | Descriptor: | Dipeptide transport ATP-binding protein DppD, Dipeptide transport ATP-binding protein DppF, Dipeptide transport system permease protein DppB, ... | | Authors: | Li, P, Huang, Y. | | Deposit date: | 2024-04-12 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural characterization of the ABC transporter DppABCDF in Escherichia coli reveals insights into dipeptide acquisition.
Plos Biol., 23, 2025
|
|
8Z1X
 
 | | Cryo-EM structure of Escherichia coli DppBCDF complex bound to AMPPNP | | Descriptor: | Dipeptide transport ATP-binding protein DppD, Dipeptide transport ATP-binding protein DppF, Dipeptide transport system permease protein DppB, ... | | Authors: | Li, P, Huang, Y. | | Deposit date: | 2024-04-12 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural characterization of the ABC transporter DppABCDF in Escherichia coli reveals insights into dipeptide acquisition.
Plos Biol., 23, 2025
|
|
4LEW
 
 | | Structure of human cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
4LEY
 
 | | Structure of mouse cGAS bound to 18 bp DNA | | Descriptor: | 18 bp dsDNA, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
3EQT
 
 | |
1J4H
 
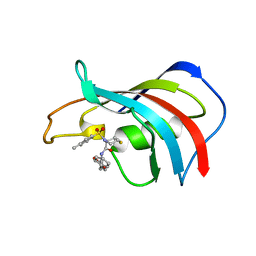 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | Descriptor: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
1J4I
 
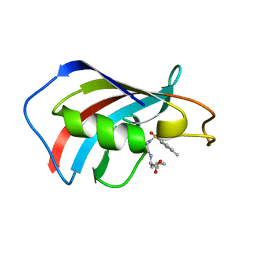 | | crystal structure analysis of the FKBP12 complexed with 000308 small molecule | | Descriptor: | 4-METHYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PENTANOIC ACID, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
8Q8P
 
 | |
8Q8Q
 
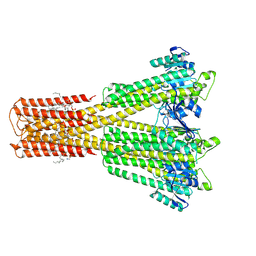 | | Cryo-EM structure of the magnesium channel CtMrs2 in the open state | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, MAGNESIUM ION, Magnesium channel Mrs2 | | Authors: | Gourdon, P, Li, P. | | Deposit date: | 2023-08-18 | | Release date: | 2024-11-27 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Closed and open structures of the eukaryotic magnesium channel Mrs2 reveal the auto-ligand-gating regulation mechanism.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6O8B
 
 | | Crystal structure of STING CTD in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
7JFM
 
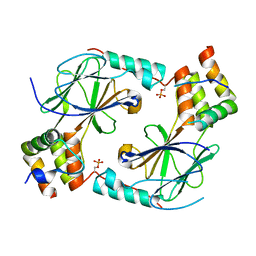 | |
7JFL
 
 | |
7EOD
 
 | | MITF HLHLZ Delta AKE | | Descriptor: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | Authors: | Li, P, Liu, Z, Fang, P, Wang, J. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
3E3H
 
 | | Crystal structure of the OP hydrolase mutant from Brevundimonas diminuta | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHYLBENZYLPHOSPHONATE, Parathion hydrolase | | Authors: | Li, P, Reeves, T.E, Grimsley, J.K, Wild, J.R. | | Deposit date: | 2008-08-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Balancing the stability and the catalytic specificities of OP hydrolases with enhanced V-agent activities.
Protein Eng.Des.Sel., 21, 2008
|
|
6O8C
 
 | | Crystal structure of STING CTT in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
5JLY
 
 | | Structure of Peroxiredoxin-1 from Schistosoma japonicum | | Descriptor: | Thioredoxin peroxidase-1 | | Authors: | Wu, Q, Huang, F, Zeng, D, Liu, X, Zhao, J, Wang, H, Peng, Y, Li, P, Li, Y. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Crystal structure of Peroxiredoxin-1 from Schistosoma japonicum
To Be Published
|
|
9J61
 
 | | Crystal structure of a cyclodipeptide synthase from Streptomyces sapporonensis | | Descriptor: | 1,2-ETHANEDIOL, Cyclodipeptide synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, P, Ren, Y, He, J, Wu, S, Wang, J, Tang, G, Fang, P. | | Deposit date: | 2024-08-14 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mutagenesis of an XYP Subfamily Cyclodipeptide Synthase Reveal Key Determinants of Enzyme Activity and Substrate Specificity.
Biochemistry, 63, 2024
|
|
4LEV
 
 | | Structure of human cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
4LEZ
 
 | | Structure of mouse cGAS bound to an 18bp DNA and cGAS product | | Descriptor: | 18bp dsDNA, Cyclic GMP-AMP synthase, ZINC ION, ... | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
