2VLD
 
 | | crystal structure of a repair endonuclease from Pyrococcus abyssi | | Descriptor: | Endonuclease NucS | | Authors: | Ren, B, Kuhn, J, Meslet-Cladiere, L, Briffotaux, J, Norais, C, Lavigne, R, Flament, D, Ladenstein, R, Myllykallio, H. | | Deposit date: | 2008-01-14 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of a novel endonuclease acting on branched DNA substrates.
EMBO J., 28, 2009
|
|
4B0N
 
 | | Crystal structure of PKS-I from the brown algae Ectocarpus siliculosus | | Descriptor: | ARACHIDONIC ACID, MALONIC ACID, POLYKETIDE SYNTHASE III | | Authors: | Leroux, C, Meslet-Cladiere, L, Delage, L, Goulitquer, S, Leblanc, C, Ar Gall, E, Stiger-Pouvreau, V, Potin, P, Czjzek, M. | | Deposit date: | 2012-07-03 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure/Function Analysis of a Type III Polyketide Synthase in the Brown Alga Ectocarpus Siliculosus Reveals a Biochemical Pathway in Phlorotannin Monomer Biosynthesis.
Plant Cell, 25, 2013
|
|
2VUG
 
 | | The structure of an archaeal homodimeric RNA ligase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PAB1020, ... | | Authors: | Brooks, M.A, Meslet-Cladiere, L, Graille, M, Kuhn, J, Blondeau, K, Myllykallio, H, van Tilbeurgh, H. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of an archaeal homodimeric ligase which has RNA circularization activity.
Protein Sci., 17, 2008
|
|
1T9H
 
 | | The crystal structure of YloQ, a circularly permuted GTPase. | | Descriptor: | ACETATE ION, CALCIUM ION, Probable GTPase engC, ... | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Cladiere, L, Antson, A.A, Isupov, M.N, Seror, S.J, Wilkinson, A.J. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of YloQ, a Circularly Permuted GTPase Essential for Bacillus Subtilis Viability.
J.Mol.Biol., 340, 2004
|
|
2W9M
 
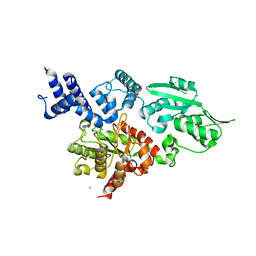 | | Structure of family X DNA polymerase from Deinococcus radiodurans | | Descriptor: | MERCURY (II) ION, POLYMERASE X, ZINC ION | | Authors: | Leulliot, N, Cladiere, L, Lecointe, F, Durand, D, Hubscher, U, van Tilbeurgh, H. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The Family X DNA Polymerase from Deinococcus Radioduran Adopts a Non-Standard Extended Conformation.
J.Biol.Chem., 284, 2009
|
|
1N45
 
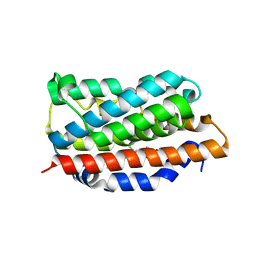 | | X-RAY CRYSTAL STRUCTURE OF HUMAN HEME OXYGENASE-1 (HO-1) IN COMPLEX WITH ITS SUBSTRATE HEME | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, heme oxygenase 1 | | Authors: | Schuller, D.J, Wilks, A, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2002-10-30 | | Release date: | 2002-11-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparison of the heme-free and -bound crystal structures of human heme oxygenase-1.
J. Biol. Chem., 278, 2003
|
|
4XE0
 
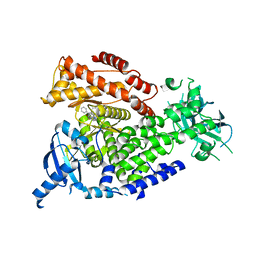 | | Idelalisib bound to the p110 subunit of PI3K delta | | Descriptor: | 5-fluoro-3-phenyl-2-[(1S)-1-(7H-purin-6-ylamino)propyl]quinazolin-4(3H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A. | | Deposit date: | 2014-12-20 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.434 Å) | | Cite: | Structural, Biochemical, and Biophysical Characterization of Idelalisib Binding to Phosphoinositide 3-Kinase delta.
J.Biol.Chem., 290, 2015
|
|
5T8I
 
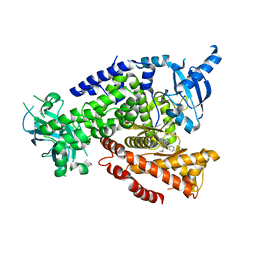 | | PI3Kdelta in complex with the inhibitor GS-9901 | | Descriptor: | 2,4-diamino-6-{[(S)-[5-chloro-8-fluoro-4-oxo-3-(pyridin-3-yl)-3,4-dihydroquinazolin-2-yl](cyclopropyl)methyl]amino}pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A. | | Deposit date: | 2016-09-07 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of GS-9901: A Potent, Selective and Metabolically Stable Inhibitor of PI3Kd
To Be Published
|
|
5T7F
 
 | | PI3Kdelta in complex with the inhibitor GS-643624 | | Descriptor: | 2,4-bis(azanyl)-6-[[(1~{S})-1-[5-chloranyl-3-(5-fluoranyl-4-methyl-pyridin-3-yl)-4-oxidanylidene-quinazolin-2-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A. | | Deposit date: | 2016-09-04 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of GS-9901: A Potent, Selective and Metabolically Stable Inhibitor of PI3Kd
To Be Published
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
6MOA
 
 | | C-terminal bromodomain of human BRD2 in complex with 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole inhibitor | | Descriptor: | 4-(2-cyclopropyl-7-(6-methylquinolin-5-yl)-1H-benzo[d]imidazol-5-yl)-3,5-dimethylisoxazole, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO7
 
 | | N-terminal bromodomain of human BRD2 with N-((4-(3-(N-cyclopentylsulfamoyl)-4-methylphenyl)-3-methylisoxazol-5-yl)methyl)acetamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-({4-[3-(cyclopentylsulfamoyl)-4-methylphenyl]-3-methyl-1,2-oxazol-5-yl}methyl)acetamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO9
 
 | | N-terminal bromodomain of human BRD2 in complex with N-cyclopentyl-7-(3,5-dimethylisoxazol-4-yl)quinoline-5-sulfonamide inhibitor | | Descriptor: | Bromodomain-containing protein 2, N-cyclopentyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)quinoline-5-sulfonamide | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
6MO8
 
 | | N-terminal bromodomain of human BRD2 in complex with 4,4'-(quinoline-5,7-diyl)bis(3,5-dimethylisoxazole) inhibitor | | Descriptor: | 5,7-bis(3,5-dimethyl-1,2-oxazol-4-yl)quinoline, Bromodomain-containing protein 2, SULFATE ION | | Authors: | Lansdon, E.B, Newby, Z.E.R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor.
Bioorg. Med. Chem., 27, 2019
|
|
4USZ
 
 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2014-07-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
4CIT
 
 | | Crystal structure of the first bacterial vanadium dependant iodoperoxidase | | Descriptor: | SODIUM ION, VANADATE ION, VANADIUM-DEPENDENT HALOPEROXIDASE | | Authors: | Rebuffet, E, Delage, L, Fournier, J.B, Rzonca, J, Potin, P, Michel, G, Czjzek, M, Leblanc, C. | | Deposit date: | 2013-12-16 | | Release date: | 2014-10-08 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Bacterial Vanadium Iodoperoxidase from the Marine Flavobacteriaceae Zobellia Galactanivorans Reveals Novel Molecular and Evolutionary Features of Halide Specificity in This Enzyme Family.
Appl.Environ.Microbiol., 80, 2014
|
|
4MNO
 
 | |
4MO0
 
 | |
5BVR
 
 | |
8R4U
 
 | | Structure of salt-inducible kinase 3 with inhibitors | | Descriptor: | 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-fluoranylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, SULFATE ION, Serine/threonine-protein kinase SIK3, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
8R4Q
 
 | | Salt inducible kinase 3 in complex with inhibitor | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, SULFATE ION, Serine/threonine-protein kinase SIK3, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.838 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
8R4V
 
 | | Structure of Salt-inducible kinase 3 in complex with inhibitor | | Descriptor: | 1-(2,4-dimethoxyphenyl)-3-(2,6-dimethylphenyl)-1-[6-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]urea, Serine/threonine-protein kinase SIK3 | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
8R4O
 
 | | Salt inducible kinase 3 in complex with inhibitor | | Descriptor: | 2-[bis(fluoranyl)methoxy]-4-[6-(2-cyanopropan-2-yl)pyrazolo[1,5-a]pyridin-3-yl]-~{N}-[(1~{R},2~{S})-2-fluoranylcyclopropyl]-6-methoxy-benzamide, SULFATE ION, Serine/threonine-protein kinase SIK3, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
2JBH
 
 | | Human phosphoribosyl transferase domain containing 1 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Welin, M, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Eklund, H, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of the Human Phosphoribosyltransferase Domain Containing Protein 1.
FEBS J., 277, 2010
|
|
5JB3
 
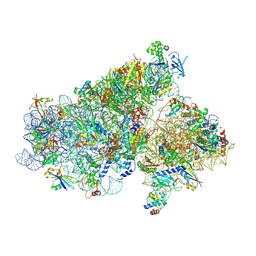 | | Cryo-EM structure of a full archaeal ribosomal translation initiation complex in the P-REMOTE conformation | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Schmitt, E, Mechulam, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5.34 Å) | | Cite: | Cryo-EM study of start codon selection during archaeal translation initiation.
Nat Commun, 7, 2016
|
|
