4KYR
 
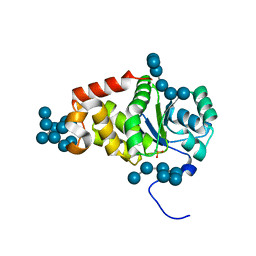 | | Structure of a product bound plant phosphatase | | Descriptor: | PHOSPHATE ION, Phosphoglucan phosphatase LSF2, chloroplastic, ... | | Authors: | Meekins, D.A, Guo, H.-F, Husodo, S, Paasch, B.C, Bridges, T.M, Santelia, D, Kotting, O, Vander Kooi, C.W, Gentry, M.S. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis Glucan Phosphatase LIKE SEX FOUR2 Reveals a Unique Mechanism for Starch Dephosphorylation.
Plant Cell, 25, 2013
|
|
4KYQ
 
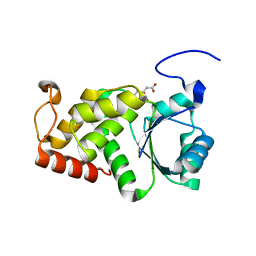 | | Structure of a product bound plant phosphatase | | Descriptor: | CITRATE ANION, Phosphoglucan phosphatase LSF2, chloroplastic | | Authors: | Meekins, D.A, Guo, H.-F, Husodo, S, Paasch, B.C, Bridges, T.M, Santelia, D, Kotting, O, Vander Kooi, C.W, Gentry, M.S. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the Arabidopsis Glucan Phosphatase LIKE SEX FOUR2 Reveals a Unique Mechanism for Starch Dephosphorylation.
Plant Cell, 25, 2013
|
|
1N87
 
 | |
4QDQ
 
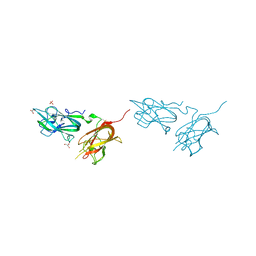 | | Physical basis for Nrp2 ligand binding | | Descriptor: | GLYCEROL, Neuropilin-2, SULFATE ION | | Authors: | Parker, M.W, Vander Kooi, C.W. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for VEGF-C Binding to Neuropilin-2 and Sequestration by a Soluble Splice Form.
Structure, 23, 2015
|
|
4PYH
 
 | |
1MNS
 
 | |
7YZY
 
 | | pMMO structure from native membranes by cryoET and STA | | Descriptor: | Methane monooxygenase subunit C2, Particulate methane monooxygenase alpha subunit, Particulate methane monooxygenase beta subunit | | Authors: | Zhu, Y, Ni, T, Zhang, P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and activity of particulate methane monooxygenase arrays in methanotrophs.
Nat Commun, 13, 2022
|
|
6UJO
 
 | |
6UK2
 
 | |
6UK4
 
 | |
6UJQ
 
 | |
7UWU
 
 | | Starch adherence system protein 6 (Sas6) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Starch Adherence System protein 6 (Sas6), ... | | Authors: | Photenhauer, A.L, Koropatkin, N.M. | | Deposit date: | 2022-05-04 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Ruminococcus bromii amylosome protein Sas6 binds single and double helical alpha-glucan structures in starch.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7UWW
 
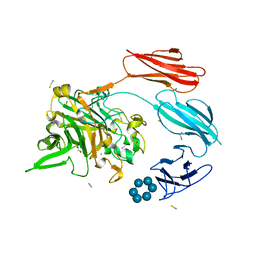 | | Sas6 with alpha-cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Photenhauer, A.L, Koropatkin, N.M. | | Deposit date: | 2022-05-04 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Ruminococcus bromii amylosome protein Sas6 binds single and double helical alpha-glucan structures in starch.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7UWV
 
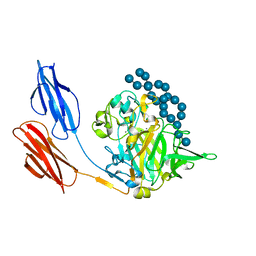 | |
4L3E
 
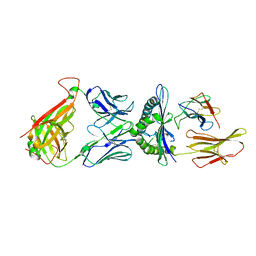 | |
6D78
 
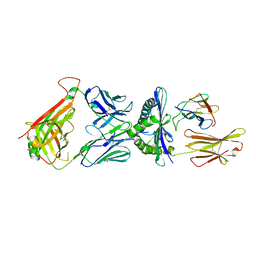 | | The complex between high-affinity TCR DMF5(alpha-D26Y,beta-L98W) and human Class I MHC HLA-A2 with the bound MART-1(27-35)peptide | | Descriptor: | Beta-2-microglobulin, DMF5 alpha chain,DMF5 alpha chain, DMF5 beta chain,DMF5 beta chain, ... | | Authors: | Hellman, L.M, Singh, N.K. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Improving T Cell Receptor On-Target Specificity via Structure-Guided Design.
Mol. Ther., 27, 2019
|
|
6DKP
 
 | | The complex among DMF5(alpha-D26Y, alpha-Y50A,beta-L98W) TCR, human Class I MHC HLA-A2 and MART-1(26-35)(A27L) peptide | | Descriptor: | Beta-2-microglobulin, DMF5 T-cell Receptor Alpha Chain fusion, DMF5 T-cell Receptor Beta Chain fusion, ... | | Authors: | Hellman, L.M, Singh, N.K. | | Deposit date: | 2018-05-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.966 Å) | | Cite: | Improving T Cell Receptor On-Target Specificity via Structure-Guided Design.
Mol. Ther., 27, 2019
|
|
5E9D
 
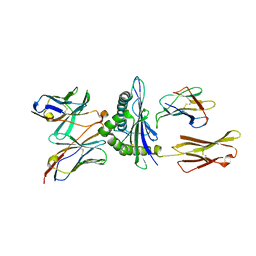 | |
5JZI
 
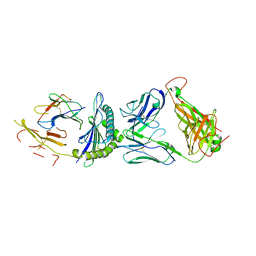 | | Crystal structure of 1406 TCR bound to HLA-A2 with HCV 1406-1415 antigen peptide | | Descriptor: | Beta-2-microglobulin, HCV1406 TCR alpha chain, HCV1406 TCR beta chain, ... | | Authors: | Wang, Y, Piepenbrink, K.H, Baker, B.M. | | Deposit date: | 2016-05-16 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How an alloreactive T-cell receptor achieves peptide and MHC specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2XR9
 
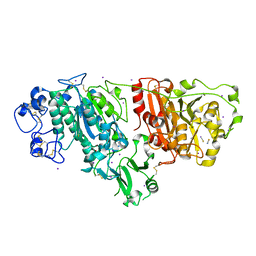 | | Crystal structure of Autotaxin (ENPP2) | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Kamtekar, S, Hausmann, J, Day, J.E, Christodoulou, E, Perrakis, A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate discrimination and integrin binding by autotaxin.
Nat. Struct. Mol. Biol., 18, 2011
|
|
2XRG
 
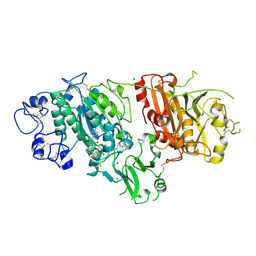 | | Crystal structure of Autotaxin (ENPP2) in complex with the HA155 boronic acid inhibitor | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Hausmann, J, Albers, H.M.H.G, Perrakis, A. | | Deposit date: | 2010-09-14 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Substrate Discrimination and Integrin Binding by Autotaxin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
