4RC5
 
 | |
5ZBZ
 
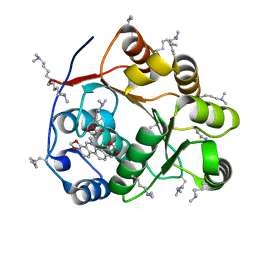 | | Crystal structure of the DEAD domain of Human eIF4A with sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Eukaryotic initiation factor 4A-I, MALONATE ION | | Authors: | Ding, Y, Ding, L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.30860257 Å) | | Cite: | Targeting the N Terminus of eIF4AI for Inhibition of Its Catalytic Recycling.
Cell Chem Biol, 26, 2019
|
|
8JI9
 
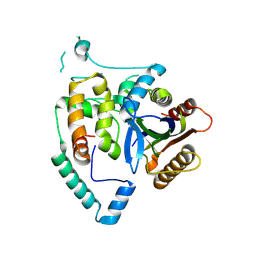 | | Crystal Structure of cas7 and AcrIE10 complex | | Descriptor: | Uncharacterized protein, type I-E Cascade subunit cas7 | | Authors: | Zhang, Y, Yu, C.Z, Sun, S.Y. | | Deposit date: | 2023-05-26 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A new anti-CRISPR gene promotes the spread of drug-resistance plasmids in Klebsiella pneumoniae.
Nucleic Acids Res., 52, 2024
|
|
5Z9R
 
 | |
3PL9
 
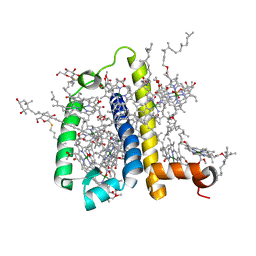 | | Crystal structure of spinach minor light-harvesting complex CP29 at 2.80 angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, M, Wan, T, Wang, L.F, Jia, C.J, Hou, Z.Q, Zhao, X.L, Zhang, J.P, Chang, W.R. | | Deposit date: | 2010-11-14 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into energy regulation of light-harvesting complex CP29 from spinach.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4RT4
 
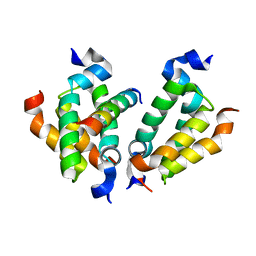 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
4RTA
 
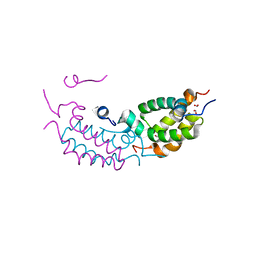 | |
5Y36
 
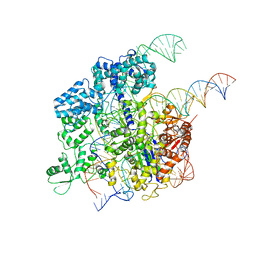 | | Cryo-EM structure of SpCas9-sgRNA-DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, complementary DNA strand, ... | | Authors: | Huang, Q, Li, G, Huai, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural insights into DNA cleavage activation of CRISPR-Cas9 system
Nat Commun, 8, 2017
|
|
6JZZ
 
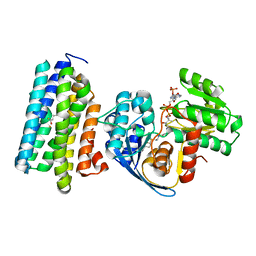 | | The crystal structure of AAR-C294S in complex with ADO. | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZY
 
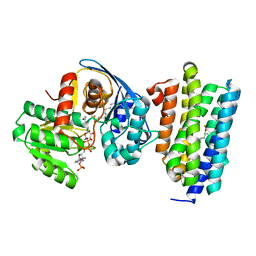 | | Crystal structure of AAR with NADPH and stearyl in complex with ADO binding a long chain carbohydrate | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZU
 
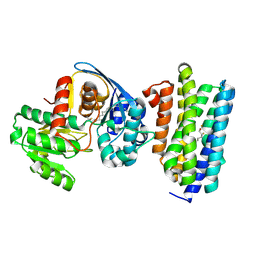 | | The crystal structure of acyl-acyl carrier protein (acyl-ACP) reductase (AAR) in complex with aldehyde deformylating oxygenase (ADO) | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZQ
 
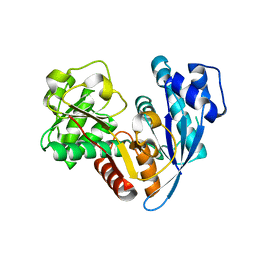 | |
7X9G
 
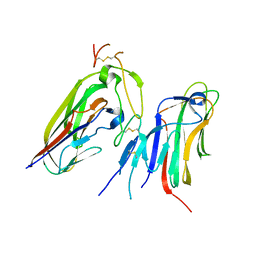 | | Crystal structure of human EDA and EDAR | | Descriptor: | Ectodysplasin-A, secreted form, Tumor necrosis factor receptor superfamily member EDAR | | Authors: | Yu, K, Wan, F, Huang, C, Wu, J, Lei, M. | | Deposit date: | 2022-03-15 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into pathogenic mechanism of hypohidrotic ectodermal dysplasia caused by ectodysplasin A variants.
Nat Commun, 14, 2023
|
|
8U1D
 
 | | Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env Local Refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1285 Heavy Chain, DH1285 Light Chain, ... | | Authors: | Thakur, B, Stalls, V.D, Acharya, P. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Vaccine induction of CD4-mimicking HIV-1 broadly neutralizing antibody precursors in macaques.
Cell, 187, 2024
|
|
8DTO
 
 | | Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-07-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
8DY6
 
 | | Vaccine elicited Antibody MU89+S27Y bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-08-03 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
3BXV
 
 | |
6UM5
 
 | | Cryo-EM structure of HIV-1 neutralizing antibody DH270 UCA3 in complex with CH848 10.17DT Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Henderson, R.C, Saunders, K.O, Haynes, B.F. | | Deposit date: | 2019-10-09 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Targeted selection of HIV-specific antibody mutations by engineering B cell maturation.
Science, 366, 2019
|
|
7P9O
 
 | | Structure of E.coli RlmJ in complex with a SAM analogue (CA) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]amino}-5'-deoxyadenosine, Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
7P8Q
 
 | | Structure of E.coli RlmJ in complex with an RNA conjugate (GA-SAM) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, RNA conjugate (GA-SAM), Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
7P9I
 
 | | Structure of E.coli RlmJ in complex with an RNA conjugate (GAA-SAM) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, RNA conjugate (GAA-SAM), Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
6UM6
 
 | | Cryo-EM structure of HIV-1 neutralizing antibody DH270.6 in complex with CH848 10.17DT Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Henderson, R.C, Saunder, K.O, Haynes, B.F. | | Deposit date: | 2019-10-09 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Targeted selection of HIV-specific antibody mutations by engineering B cell maturation.
Science, 366, 2019
|
|
6UM7
 
 | | Cryo-EM structure of vaccine-elicited HIV-1 neutralizing antibody DH270.mu1 in complex with CH848 10.17DT Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH270.mu1 Fab Heavy Chain, DH270.mu1 Fab Light chain, ... | | Authors: | Acharya, P, Henderson, R.C, Saunders, K, Haynes, B.F. | | Deposit date: | 2019-10-09 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeted selection of HIV-specific antibody mutations by engineering B cell maturation.
Science, 366, 2019
|
|
6S73
 
 | | Crystal structure of Nek7 SRS mutant bound to compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, Serine/threonine-protein kinase Nek7 | | Authors: | Nasir, N, Byrne, M.J, Bhatia, C, Bayliss, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
6S75
 
 | | Crystal structure of Nek7 bound to compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, Serine/threonine-protein kinase Nek7 | | Authors: | Nasir, N, Bayliss, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
