7AH8
 
 | | NF-Y bound to suramin inhibitor | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, CITRATE ANION, GLYCEROL, ... | | Authors: | Nardone, V, Chaves-Sanjuan, A, Lapi, M, Nardini, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.70001364 Å) | | Cite: | Structural Basis of Inhibition of the Pioneer Transcription Factor NF-Y by Suramin.
Cells, 9, 2020
|
|
7QQE
 
 | | Nuclear factor one X - NFIX in P41212 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nuclear factor 1 X-type, ZINC ION | | Authors: | Lapi, M, Chaves-Sanjuan, A, Gourlay, L.J, Tiberi, M, Polentarutti, M, Demitri, N, Bais, G, Nardini, M. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nuclear factor one X - NFIX in P41212
To Be Published
|
|
7QQD
 
 | | Nuclear factor one X - NFIX in P21 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NFI binding site (forward), NFI binding site (reverse), ... | | Authors: | Lapi, M, Chaves-Sanjuan, A, Gourlay, L.J, Tiberi, M, Polentarutti, M, Demitri, N, Bais, G, Nardini, M. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nuclear factor one X - NFIX in P41212
To Be Published
|
|
8CPE
 
 | |
7NP4
 
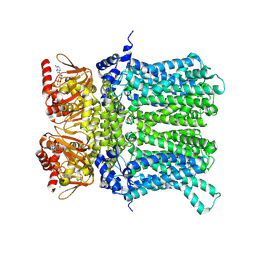 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP3
 
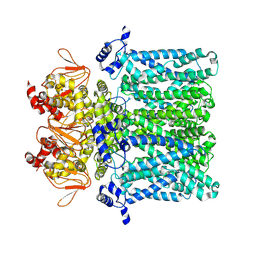 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
5FYX
 
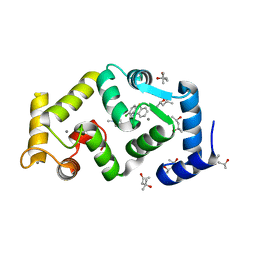 | | Crystal structure of Drosophila NCS-1 bound to penothiazine FD16 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, FREQUENIN 2, ... | | Authors: | Martinez-Gonzalez, L, Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NWI
 
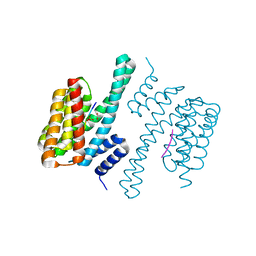 | | 14-3-3c in complex with CPP | | Descriptor: | 14-3-3 c-1 protein, ACETATE ION, Potassium channel KAT1 | | Authors: | Saponaro, A, Porro, A, Chaves-Sanjuan, A, Nardini, M, Thiel, G, Moroni, A. | | Deposit date: | 2017-05-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fusicoccin Activates KAT1 Channels by Stabilizing Their Interaction with 14-3-3 Proteins.
Plant Cell, 29, 2017
|
|
5NWK
 
 | | 14-3-3c in complex with CPP and fusicoccin | | Descriptor: | 14-3-3 c-1 protein, FUSICOCCIN, PHOSPHOSERINE, ... | | Authors: | Saponaro, A, Porro, A, Chaves-Sanjuan, A, Nardini, M, Thiel, G, Moroni, A. | | Deposit date: | 2017-05-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fusicoccin Activates KAT1 Channels by Stabilizing Their Interaction with 14-3-3 Proteins.
Plant Cell, 29, 2017
|
|
5NWJ
 
 | | 14-3-3c in complex with CPP7 | | Descriptor: | 14-3-3-like protein C, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saponaro, A, Porro, A, Chaves-Sanjuan, A, Nardini, M, Thiel, G, Moroni, A. | | Deposit date: | 2017-05-06 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Fusicoccin Activates KAT1 Channels by Stabilizing Their Interaction with 14-3-3 Proteins.
Plant Cell, 29, 2017
|
|
5G49
 
 | | Crystal structure of the Arabodopsis thaliana histone-fold dimer L1L NF-YC3 | | Descriptor: | ACETATE ION, CALCIUM ION, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT B-6, ... | | Authors: | Gnesutta, N, Saad, D, Chaves-Sanjuan, A, Mantovani, R, Nardini, M. | | Deposit date: | 2016-05-06 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Arabidopsis thaliana L1L/NF-YC3 Histone-fold Dimer Reveals Specificities of the LEC1 Family of NF-Y Subunits in Plants.
Mol Plant, 10, 2017
|
|
9FAA
 
 | | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
9FAB
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph
To be published
|
|
9FAC
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - mixed polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
4BY4
 
 | |
4BY5
 
 | |
8A11
 
 | | Cryo-EM structure of the Human SHMT1-RNA complex | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 2024
|
|
7ZIR
 
 | | Cryo-EM structure of hnRNPDL amyloid fibrils | | Descriptor: | Heterogeneous nuclear ribonucleoprotein D-like | | Authors: | Garcia-Pardo, J, Chaves-Sanjuan, A, Bartolome-Nafria, A, Gil-Garcia, M, Visentin, C, Bolognesi, M, Ricagno, S, Ventura, S. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of hnRNPDL-2 fibrils, a functional amyloid associated with limb-girdle muscular dystrophy D3.
Nat Commun, 14, 2023
|
|
6RJD
 
 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA59-ntPAM complex. | | Descriptor: | Streptococcus Thermophilus 1 Cas9, ntPAM, sgRNA (78-MER), ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJ9
 
 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA20-AcrIIA6 monomeric assembly. | | Descriptor: | AcrIIA6, CRISPR-associated endonuclease Cas9 1, sgRNA, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJA
 
 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA20-AcrIIA6 dimeric assembly. | | Descriptor: | AcrIIA6, CRISPR-associated endonuclease Cas9 1, RNA (78-MER), ... | | Authors: | Goulet, A, Cambillau, C, Chaves-Sanjuan, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
6RJG
 
 | | Cryo-EM structure of St1Cas9-sgRNA-AcrIIA6-tDNA59-ntPAM complex. | | Descriptor: | AcrIIA6, Cas 9, ntPAM, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
7ZH7
 
 | |
6EPA
 
 | | Structure of dNCS-1 bound to the NCS-1/Ric8a protein/protein interaction regulator IGS-1.76 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, FI18190p1, ... | | Authors: | Sanchez-Barrena, M.J, Daniel, M, Infantes, L. | | Deposit date: | 2017-10-11 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Deciphering the Inhibition of the Neuronal Calcium Sensor 1 and the Guanine Exchange Factor Ric8a with a Small Phenothiazine Molecule for the Rational Generation of Therapeutic Synapse Function Regulators.
J. Med. Chem., 61, 2018
|
|
7PQ9
 
 | | Crystal structure of Bacillus clausii pdxR at 2.8 Angstroms resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vivoli Vega, M, Isupov, M.N, Harmer, N. | | Deposit date: | 2021-09-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
