7RXV
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine, Malonate (MLA) and MgF3 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, GLYCEROL, ... | | Authors: | Fedorov, E, Niland, C.N, Schramm, V.L, Ghosh, A. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Mechanism of Triphosphate Hydrolysis by Human MAT2A at 1.07 angstrom Resolution.
J.Am.Chem.Soc., 143, 2021
|
|
2A0X
 
 | | Structure of human purine nucleoside phosphorylase H257F mutant | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Murkin, A.S, Shi, W, Schramm, V.L. | | Deposit date: | 2005-06-17 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
Biochemistry, 46, 2007
|
|
7RXW
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and inhibitor imido-diphosphate (PNP) | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, ... | | Authors: | Fedorov, E, Niland, C.N, Schramm, V.L, Ghosh, A. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Mechanism of Triphosphate Hydrolysis by Human MAT2A at 1.07 angstrom Resolution.
J.Am.Chem.Soc., 143, 2021
|
|
7RXX
 
 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and two sulfate in the active site | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Fedorov, E, Niland, C.N, Schramm, V.L, Ghosh, A. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism of Triphosphate Hydrolysis by Human MAT2A at 1.07 angstrom Resolution.
J.Am.Chem.Soc., 143, 2021
|
|
1B8N
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Strokopytov, B.V, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic dissection of the hydrogen bond network for transition-state analogue binding to purine nucleoside phosphorylase
Biochemistry, 41, 2002
|
|
1I80
 
 | | CRYSTAL STRUCTURE OF M. TUBERCULOSIS PNP IN COMPLEX WITH IMINORIBITOL, 9-DEAZAHYPOXANTHINE AND PHOSPHATE ION | | Descriptor: | 9-DEAZAHYPOXANTHINE, IMINORIBITOL, PHOSPHATE ION, ... | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2001-03-12 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
1BZY
 
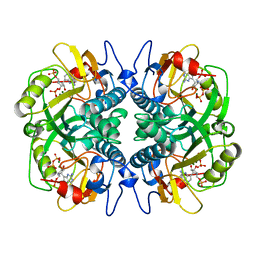 | | HUMAN HGPRTASE WITH TRANSITION STATE INHIBITOR | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER, ... | | Authors: | Shi, W, Li, C, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1998-11-05 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor.
Nat.Struct.Biol., 6, 1999
|
|
1CJB
 
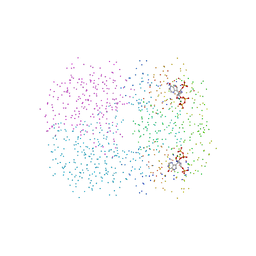 | | MALARIAL PURINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, MAGNESIUM ION, PROTEIN (HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE), ... | | Authors: | Shi, W, Li, C.M, Tyler, P.C, Furneaux, R.H, Cahill, S.M, Girvin, M.E, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-04-08 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of malarial purine phosphoribosyltransferase in complex with a transition-state analogue inhibitor.
Biochemistry, 38, 1999
|
|
1L1Q
 
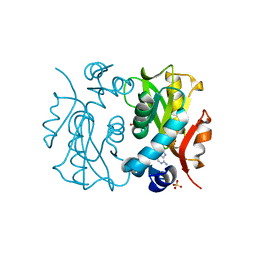 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine | | Descriptor: | 9-DEAZAADENINE, Adenine phosphoribosyltransferase, SULFATE ION | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
1L1R
 
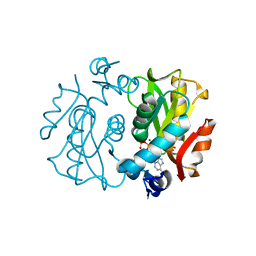 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine, Mg2+ and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAADENINE, Adenine phosphoribosyltransferase, ... | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
2ISC
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with DADMe-Imm-A | | Descriptor: | (3R,4R)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-(HYDROXYMETHYL)PYRROLIDIN-3-OL, PHOSPHATE ION, purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2006-10-17 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues
Biochemistry, 46, 2007
|
|
2I4T
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with Imm-A | | Descriptor: | 3,4-PYRROLIDINEDIOL,2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)-2S,3S,4R,5R, PHOSPHATE ION, Trichomonas vaginalis purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Schramm, V.L, Almo, S.C. | | Deposit date: | 2006-08-22 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues.
Biochemistry, 46, 2007
|
|
5TC5
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with butylthio-DADMe-Immucillin-A and chloride | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, CHLORIDE ION, S-methyl-5'-thioadenosine phosphorylase | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | TBA
To be published
|
|
5TC8
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | TBA
To be published
|
|
5TC6
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with propylthio-immucillin-A | | Descriptor: | (2S,3S,4R,5S)-2-(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)-5-[(propylsulfanyl)methyl]pyrrolidine-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | TBA
To be published
|
|
5TC7
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with 5'-methylthiotubercidin at 1.75 angstrom | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | TBA
To be published
|
|
4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
1Y6Q
 
 | | Cyrstal structure of MTA/AdoHcy nucleosidase complexed with MT-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
1ZOS
 
 | | Structure of 5'-methylthionadenosine/S-Adenosylhomocysteine nucleosidase from S. pneumoniae with a transition-state inhibitor MT-ImmA | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5'-methylthioadenosine / S-adenosylhomocysteine nucleosidase | | Authors: | Shi, W, Singh, V, Zhen, R, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2005-05-13 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and inhibition of a quorum sensing target from Streptococcus pneumoniae.
Biochemistry, 45, 2006
|
|
6AQU
 
 | | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase: The M183L mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition inPlasmodium falciparum.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AQS
 
 | | Crystal structure of Plasmodium falciparum purine nucleoside phosphorylase (V181D) mutant complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition in
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AYR
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYT
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with pyrazinylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(pyrazin-2-ylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYO
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 5'-deoxy-5'-Propyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-propylpyrrolidin-3-ol, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYM
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) | | Descriptor: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
