8JPN
 
 | | Cryo-EM structure of succinate receptor bound to cis-epoxysuccinic acid coupling to Gi | | Descriptor: | (2R,3S)-oxirane-2,3-dicarboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-06-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
5XOB
 
 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
3SX0
 
 | | Crystal structure of Dot1l in complex with a brominated SAH analog | | Descriptor: | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoic acid (non-preferred name), Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-14 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Bromo-deaza-SAH: a potent and selective DOT1L inhibitor.
Bioorg. Med. Chem., 21, 2013
|
|
2Z60
 
 | | Crystal Structure of the T315I Mutant of Abl kinase bound with PPY-A | | Descriptor: | 5-[3-(2-METHOXYPHENYL)-1H-PYRROLO[2,3-B]PYRIDIN-5-YL]-N,N-DIMETHYLPYRIDINE-3-CARBOXAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Dalgarno, D, Zhu, X. | | Deposit date: | 2007-07-21 | | Release date: | 2007-09-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the T315I Mutant of Abl Kinase
Chem.Biol.Drug Des., 70, 2007
|
|
8XJ4
 
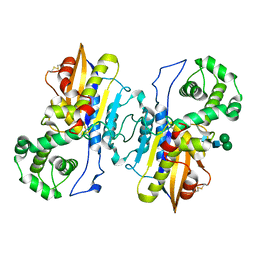 | | Structure of prostatic acid phosphatase in human semen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prostatic acid phosphatase, alpha-D-mannopyranose, ... | | Authors: | Liu, X.Z, Li, J.L, Deng, D, Wang, X. | | Deposit date: | 2023-12-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Purification, identification and Cryo-EM structure of prostatic acid phosphatase in human semen.
Biochem.Biophys.Res.Commun., 702, 2024
|
|
7ESI
 
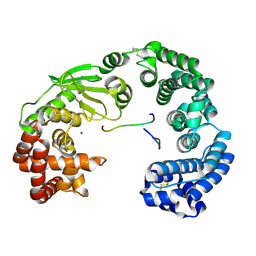 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 at 1. 8 angstrom resolution. | | Descriptor: | CALCIUM ION, Collagenase unit (CU), Peptide P1, ... | | Authors: | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-09 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun, 13, 2022
|
|
7CQQ
 
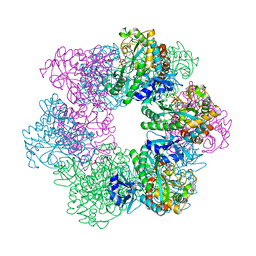 | | GmaS in complex with AMPPNP and MetSox | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQL
 
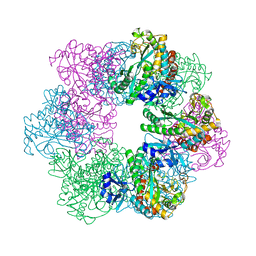 | | Apo GmaS without ligand | | Descriptor: | Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
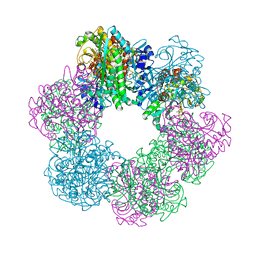 | | GmaS in complex with AMPPCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
6LJA
 
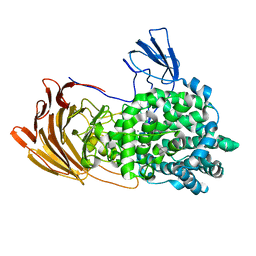 | | Crystal Structure of exoHep from Bacteroides intestinalis DSM 17393 complexed with disaccharide product | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
7CQX
 
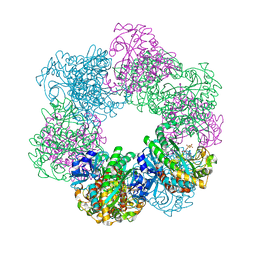 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQW
 
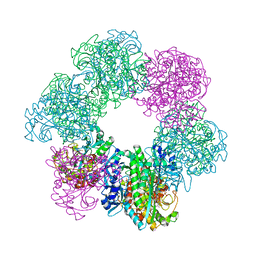 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
6LJL
 
 | | Crystal Structure of exoHep-Y390A/H555A complexed with a tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
7CM9
 
 | | DMSP lyase DddX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DMSP lyase, SULFATE ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-07-25 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.
Elife, 10, 2021
|
|
8HIH
 
 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Shi, J, Xu, J.C. | | Deposit date: | 2022-11-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7EVR
 
 | | Crystal structure of hnRNP L RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L, SHI domain from Histone-lysine N-methyltransferase SETD2 | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
7EVS
 
 | | Crystal structure of hnRNP LL RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L-like, SHI domain from Histone-lysine N-methyltransferase SETD2, SULFATE ION | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
3NY1
 
 | | Structure of the ubr-box of the UBR1 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NY3
 
 | | Structure of the ubr-box of UBR2 in complex with N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR2, N-degron, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4H12
 
 | | The crystal structure of methyltransferase domain of human SET domain-containing protein 2 in complex with S-adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Amaya, M.F, Dong, A, Zeng, H, Mackenzie, F, Bunnage, M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Sinefungin Derivatives as Inhibitors and Structure Probes of Protein Lysine Methyltransferase SETD2.
J.Am.Chem.Soc., 134, 2012
|
|
3PHD
 
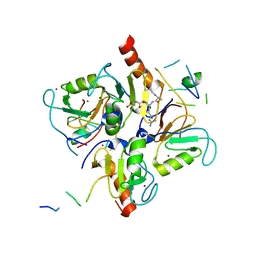 | | Crystal structure of human HDAC6 in complex with ubiquitin | | Descriptor: | Histone deacetylase 6, Polyubiquitin, ZINC ION | | Authors: | Dong, A, Qui, W, Ravichandran, M, Schuetz, A, Loppnau, P, Li, F, Mackenzie, F, Kozieradzki, I, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Aggregates Are Recruited to Aggresome by Histone Deacetylase 6 via Unanchored Ubiquitin C Termini.
J.Biol.Chem., 287, 2012
|
|
8YZK
 
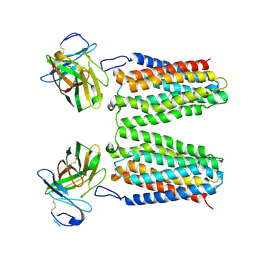 | | Orphan receptor GPRC5D in complex with scFv150-18 | | Descriptor: | Soluble cytochrome b562,G-protein coupled receptor family C group 5 member D, scFv | | Authors: | Yan, P, Lin, X, Xu, F. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The binding mechanism of an anti-multiple myeloma antibody to the human GPRC5D homodimer.
Nat Commun, 15, 2024
|
|
8ZSS
 
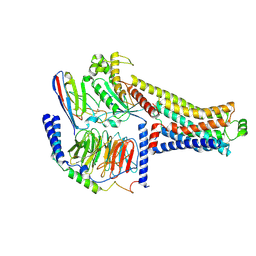 | | Cryo-EM structure of the RO5263397-bound hTAAR1-Gs complex | | Descriptor: | (4~{S})-4-(3-fluoranyl-2-methyl-phenyl)-1,3-oxazolidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
