2KKO
 
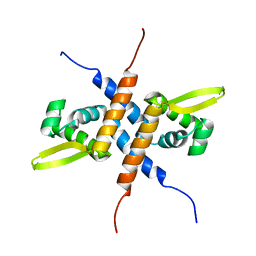 | | Solution NMR structure of the homodimeric winged helix-turn-helix DNA-binding domain (fragment 1-100) Mb0332 from Mycobacterium bovis, a possible ArsR-family transcriptional regulator. Northeast Structural Genomics Consortium Target MbR242E. | | Descriptor: | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (POSSIBLY ARSR-FAMILY) | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the homodimeric winged helix-turn-helix DNA-binding domain (fragment 1-100) Mb0332 from Mycobacterium bovis, a possible ArsR-family transcriptional regulator. Northeast Structural Genomics Consortium Target MbR242E.
To be Published
|
|
2KJ5
 
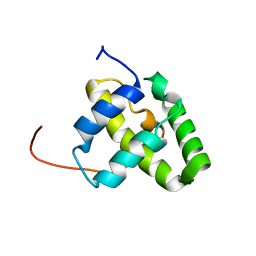 | | Solution NMR structure of a domain from a putative phage integrase protein Nmul_A0064 from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR46C | | Descriptor: | Phage integrase | | Authors: | Mills, J.L, Eletsky, A, Ghosh, A, Wang, D, Lee, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a domain from a putative phage integrase protein Nmul_A0064 from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR46C.
To be Published
|
|
6L84
 
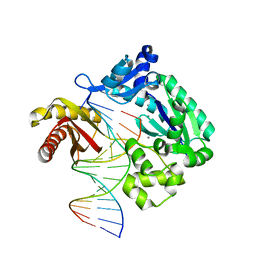 | | Complex of DNA polymerase IV and D-DNA duplex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), DNA (5'-D(P*CP*GP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Chung, H.S, An, J, Hwang, D. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The crystal structure of a natural DNA polymerase complexed with mirror DNA.
Chem.Commun.(Camb.), 56, 2020
|
|
6L97
 
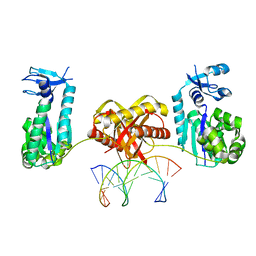 | | Complex of DNA polymerase IV and L-DNA duplex | | Descriptor: | DNA (5'-D(*(0DG)P*(0DG)P*(0DG)P*(0DG)P*(0DG)P*(0DA)P*(0DA)P*(0DG)P*(0DG)P*(0DA)P*(0DT)P*(0DT)P*(0DC)P*(0DC))-3'), DNA (5'-D(P*(0DG)P*(0DG)P*(0DA)P*(0DA)P*(0DT)P*(0DC)P*(0DC)P*(0DT)P*(0DT)P*(0DC)P*(0DC)P*(0DC)P*(0DC)P*(0DC))-3'), DNA polymerase IV | | Authors: | Chung, H.S, An, J, Hwang, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.362 Å) | | Cite: | The crystal structure of a natural DNA polymerase complexed with mirror DNA.
Chem.Commun.(Camb.), 56, 2020
|
|
8JED
 
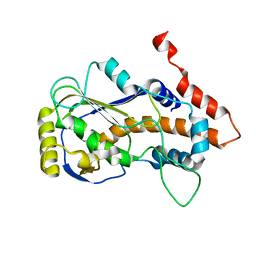 | | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors | | Descriptor: | mRNA-capping enzyme regulatory subunit OPG124 | | Authors: | Wang, D, Zhao, R, Shu, W, Hu, W, Wang, M, Cao, J, Zhou, X. | | Deposit date: | 2023-05-15 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors.
Int.J.Biol.Macromol., 253, 2023
|
|
7XUX
 
 | |
9BFL
 
 | |
8FB9
 
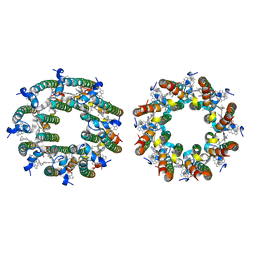 | | LH2-LH3 antenna in anti parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-11-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FBB
 
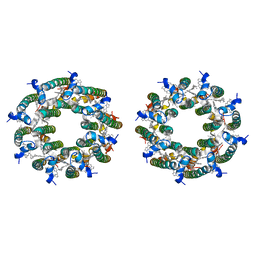 | | LH2-LH3 antenna in parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-11-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7LIP
 
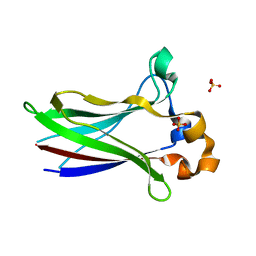 | | X-ray structure of SPOP MATH domain (D140G) | | Descriptor: | SULFATE ION, Speckle-type POZ protein | | Authors: | Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | ATM-phosphorylated SPOP contributes to 53BP1 exclusion from chromatin during DNA replication.
Sci Adv, 7, 2021
|
|
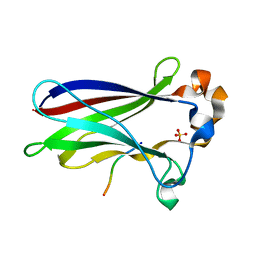
7LIN
 
 | |
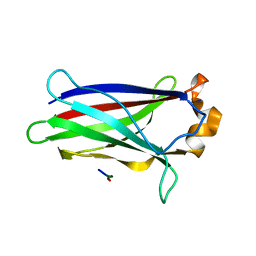
7LIO
 
 | |
7LIQ
 
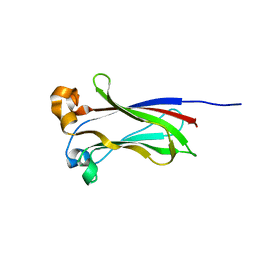 | |
8X5X
 
 | |
8X63
 
 | |
8X64
 
 | |
8X5Y
 
 | |
6B0R
 
 | |
6BLW
 
 | |
7TV3
 
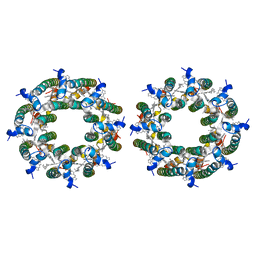 | | LH2-LH3 antenna in parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (11.4 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TUW
 
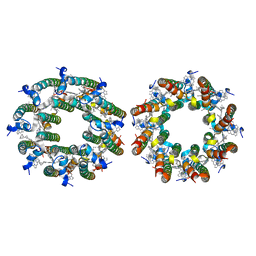 | | LH2-LH3 antenna in anti parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5GSY
 
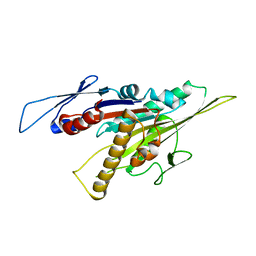 | | Kinesin-8 motor, KIF19A, in the nucleotide-free state complexed with GDP-taxol microtubule | | Descriptor: | Kinesin-like protein KIF19 | | Authors: | Morikawa, M, Nitta, R, Yajima, H, Shigematsu, H, Kikkawa, M, Hirokawa, N. | | Deposit date: | 2016-08-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
6B0O
 
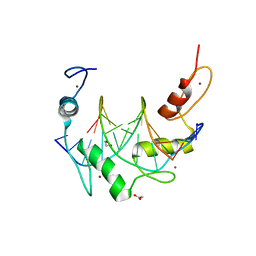 | | Zinc finger Domain of WT1(-KTS form) with 12+1mer Oligonucleotide with 3' Triplet TGT | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*TP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-09-14 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Role for first zinc finger of WT1 in DNA sequence specificity: Denys-Drash syndrome-associated WT1 mutant in ZF1 enhances affinity for a subset of WT1 binding sites.
Nucleic Acids Res., 46, 2018
|
|
6B0P
 
 | | Zinc finger Domain of WT1(-KTS form) with 12+1mer Oligonucleotide with 3' Triplet GGT | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*AP*CP*CP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*TP*GP*GP*GP*AP*GP*GP*GP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-09-14 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Role for first zinc finger of WT1 in DNA sequence specificity: Denys-Drash syndrome-associated WT1 mutant in ZF1 enhances affinity for a subset of WT1 binding sites.
Nucleic Acids Res., 46, 2018
|
|
6B0Q
 
 | | Zinc finger Domain of WT1(-KTS form) with 13+1mer Oligonucleotide with 3' Triplet TGT | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*TP*GP*GP*GP*AP*GP*TP*GP*TP*T)-3'), SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-09-14 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Role for first zinc finger of WT1 in DNA sequence specificity: Denys-Drash syndrome-associated WT1 mutant in ZF1 enhances affinity for a subset of WT1 binding sites.
Nucleic Acids Res., 46, 2018
|
|
