8EEU
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05 | | Descriptor: | Coat protein, Fab SKT05 heavy chain, Fab SKT05 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8EEV
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20 | | Descriptor: | Coat protein, Fab SKT20 heavy chain, Fab SKT20 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8F6X
 
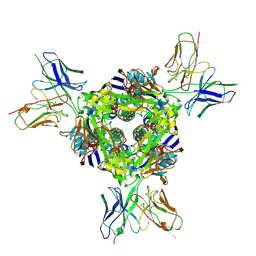 | | cryo-EM structure of a structurally designed Human metapneumovirus F protein in complex with antibody MPE8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MPE8 Single chain variable fragment, Structurally designed HMPV F protein HMPV_v3B_D12_DS454,Fibritin | | Authors: | Zhou, T, Kwong, P.D, Morano, N.C, Ou, L. | | Deposit date: | 2022-11-17 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | cryo-EM structure of a structurally designed Human metapneumovirus F protein in complex with antibody MPE8
To Be Published
|
|
8F7T
 
 | | Glycan-Base ConC Env Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env gp120, ... | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-20 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases.
Iscience, 26, 2023
|
|
8F7Z
 
 | | VRC34.01_mm28 bound to fusion peptide | | Descriptor: | HIV-1 Env Fusion Peptide, VRC34_m228 Light Chain, VRC34_mm28 Heavy Chain | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8FA0
 
 | |
8F9Z
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with NBD-14204, an HIV-1 gp120 antagonist | | Descriptor: | (5M)-N-{(1S)-2-amino-1-[5-(hydroxymethyl)-1,3-thiazol-2-yl]ethyl}-5-[5-(trifluoromethyl)pyridin-2-yl]-1H-pyrrole-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Antiviral Activity and Crystal Structures of HIV-1 gp120 Antagonists.
Int J Mol Sci, 23, 2022
|
|
8FBW
 
 | | Crystal structure of SIV-1 V2 antibody NCI05 in complex with a V2 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, T, Kwong, P.D, Van Wazer, D.J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Effect of Passive Administration of Monoclonal Antibodies Recognizing Simian Immunodeficiency Virus (SIV) V2 in CH59-Like Coil/Helical or beta-Sheet Conformations on Time of SIV mac251 Acquisition.
J.Virol., 97, 2023
|
|
8FIS
 
 | | Structure of Bispecific CAP256V2LS-J3 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-16 | | Release date: | 2023-02-01 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Bispecific antibody CAP256.J3LS targets V2-apex and CD4-binding sites with high breadth and potency.
Mabs, 15, 2023
|
|
8FR6
 
 | | Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Wang, S, Kwong, P.D. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8FLW
 
 | | Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Improved HIV-1 neutralization breadth and potency of V2-apex antibodies by in silico design.
Cell Rep, 42, 2023
|
|
8ELI
 
 | |
7L0L
 
 | | Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited, human antibody 316-310-1B11 in complex with an H2 CAN05 HA trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 316-310-1B11 Heavy Chain, 316-310-1B11 Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-11-03 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination.
Nat Med, 28, 2022
|
|
7LQV
 
 | | Cryo-EM structure of NTD-directed neutralizing antibody 4-8 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-8 Heavy Chain, 4-8 Light chain, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cell Host Microbe, 2021
|
|
7LQW
 
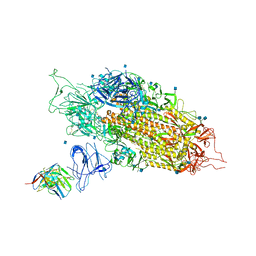 | | Cryo-EM structure of NTD-directed neutralizing antibody 2-17 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-17 Heavy Chain, 2-17 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
7LKG
 
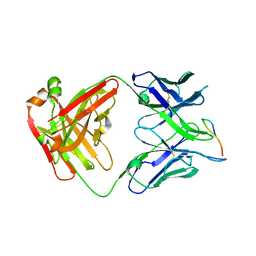 | |
7LKB
 
 | |
7LPN
 
 | |
7MFB
 
 | | Crystal structure of antibody 10E8v4 Fab - light chain H31F variant | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7MFA
 
 | | Crystal structure of antibody 10E8v4-P100fA+P100gA Fab | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7MF7
 
 | | Crystal structure of antibody 10E8v4-P100gA Fab | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7MF8
 
 | | Crystal structure of antibody 10E8v4-P100fA Fab in space group P6422 | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7MF9
 
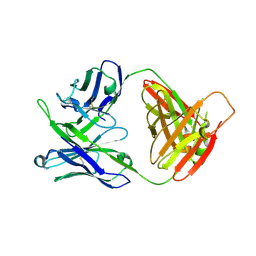 | | Crystal structure of antibody 10E8v4-P100fA Fab in space group C2 | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
7MFG
 
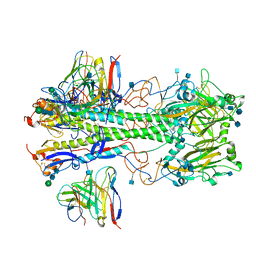 | | Cryo-EM structure of the VRC310 clinical trial, vaccine-elicited, human antibody 310-030-1D06 Fab in complex with an H1 NC99 HA trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 310-030-1D06 Heavy, 310-030-1D06 Light, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2021-04-09 | | Release date: | 2021-11-03 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination.
Nat Med, 28, 2022
|
|
7MTD
 
 | | Structure of aged SARS-CoV-2 S2P spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tsybovsky, Y, Olia, A.S, Kwong, P.D. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity.
J.Biol.Chem., 297, 2021
|
|
