1I8J
 
 | | CRYSTAL STRUCTURE OF PORPHOBILINOGEN SYNTHASE COMPLEXED WITH THE INHIBITOR 4,7-DIOXOSEBACIC ACID | | Descriptor: | 4,7-DIOXOSEBACIC ACID, MAGNESIUM ION, PORPHOBILINOGEN SYNTHASE, ... | | Authors: | Kervinen, J, Jaffe, E.K, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic basis for suicide inactivation of porphobilinogen synthase by 4,7-dioxosebacic acid, an inhibitor that shows dramatic species selectivity.
Biochemistry, 40, 2001
|
|
1DPJ
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH IA3 PEPTIDE INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEINASE A, PROTEINASE INHIBITOR IA3 PEPTIDE, ... | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-27 | | Release date: | 2000-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1DP5
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT INHIBITOR | | Descriptor: | PROTEINASE A, PROTEINASE INHIBITOR IA3, beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-23 | | Release date: | 2000-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1G3P
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAINS OF BACTERIOPHAGE MINOR COAT PROTEIN G3P | | Descriptor: | MINOR COAT PROTEIN, SULFATE ION | | Authors: | Lubkowski, J, Hennecke, F, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1997-12-22 | | Release date: | 1998-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p.
Nat.Struct.Biol., 5, 1998
|
|
1EMC
 
 | |
1QMH
 
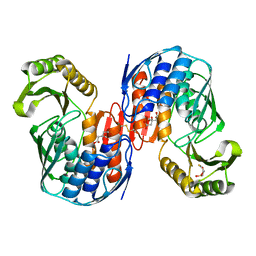 | | Crystal structure of RNA 3'-terminal phosphate cyclase, an ubiquitous enzyme with unusual topology | | Descriptor: | 1-HYDROXYSULFANYL-4-MERCAPTO-BUTANE-2,3-DIOL, CITRIC ACID, RNA 3'-TERMINAL PHOSPHATE CYCLASE | | Authors: | Palm, G.J, Billy, E, Filipowicz, W, Wlodawer, A. | | Deposit date: | 1999-09-28 | | Release date: | 2000-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of RNA 3'-Terminal Phosphate Cyclase, a Ubiquitous Enzyme with Unusual Topology
Structure, 8, 2000
|
|
1QMI
 
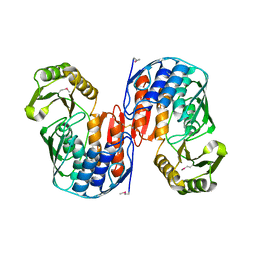 | | Crystal structure of RNA 3'-terminal phosphate cyclase, an ubiquitous enzyme with unusual topology | | Descriptor: | RNA 3'-TERMINAL PHOSPHATE CYCLASE | | Authors: | Palm, G.J, Billy, E, Filipowicz, W, Wlodawer, A. | | Deposit date: | 1999-09-28 | | Release date: | 2000-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of RNA 3'-Terminal Phosphate Cyclase, a Ubiquitous Enzyme with Unusual Topology
Structure, 8, 2000
|
|
1EMF
 
 | |
1EME
 
 | |
1CMS
 
 | |
1EML
 
 | |
1EMK
 
 | |
1EMM
 
 | |
1CZ9
 
 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN (D64N) FROM CITRATE | | Descriptor: | AVIAN SARCOMA VIRUS INTEGRASE, CITRIC ACID, SULFATE ION | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-09-01 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
1DOK
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1CXQ
 
 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN FROM AMMONIUM SULFATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE, GLYCEROL | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-08-30 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
1CZB
 
 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN FROM HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-09-01 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
1DOL
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, I-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1 | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A. | | Deposit date: | 1996-11-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1K2A
 
 | | Modified Form of Eosinophil-derived Neurotoxin | | Descriptor: | SULFATE ION, eosinophil-derived neurotoxin | | Authors: | Chang, C, Newton, D.L, Rybak, S.M, Wlodawer, A. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystallographic and functional studies of a modified form of eosinophil-derived neurotoxin (EDN) with novel biological activities.
J.Mol.Biol., 317, 2002
|
|
1HIV
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF HIV-1 PROTEASE WITH A DIHYDROETHYLENE-CONTAINING INHIBITOR: COMPARISONS WITH MOLECULAR MODELING | | Descriptor: | 4-[(2R)-3-{[(1S,2S,3R,4S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methyl-4-({(1S,2R)-2-methyl-1-[(pyridin-2-ylmethyl)carba moyl]butyl}carbamoyl)hexyl]amino}-2-{[(naphthalen-1-yloxy)acetyl]amino}-3-oxopropyl]-1H-imidazol-3-ium, HIV-1 PROTEASE | | Authors: | Thanki, N, Wlodawer, A. | | Deposit date: | 1992-02-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a complex of HIV-1 protease with a dihydroxyethylene-containing inhibitor: comparisons with molecular modeling.
Protein Sci., 1, 1992
|
|
1K6U
 
 | | Crystal Structure of Cyclic Bovine Pancreatic Trypsin Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Botos, I, Wu, Z, Lu, W, Wlodawer, A. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of a cyclic form of bovine pancreatic trypsin inhibitor.
FEBS Lett., 509, 2001
|
|
7RY7
 
 | |
1RTA
 
 | |
1RTB
 
 | |
4PGA
 
 | | GLUTAMINASE-ASPARAGINASE FROM PSEUDOMONAS 7A | | Descriptor: | AMMONIUM ION, GLUTAMINASE-ASPARAGINASE, SULFATE ION | | Authors: | Jakob, C.G, Lewinski, K, Lacount, M.W, Roberts, J, Lebioda, L. | | Deposit date: | 1997-01-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ion binding induces closed conformation in Pseudomonas 7A glutaminase-asparaginase (PGA): crystal structure of the PGA-SO4(2-)-NH4+ complex at 1.7 A resolution.
Biochemistry, 36, 1997
|
|
